W10 L1 - Overview of membrane traffic and the methods to study it
1/59
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
60 Terms
What are compartments of a eukaryotic cell defined by?
The presence of internal membrane compartments each with a specific composition and function
What are organelles?
Membrane enclosed compartments
What is an example of a membrane-less organelle?
Stress granules
ER - How much of a volume of the cell is membrane-enclosed compartments?
On average, the membrane-enclosed compartments together occupy nearly half the volume of a cell
ER - how do organelles vary in plasma cells?
plasma cells which daily secrete their own weight in antibody molecules into the bloodstream, contain vastly amplified amounts of rough ER, which is found in large, flat sheets
ER - how do organelles vary in cardiac muscle cells?
Cardiac muscle cells instead expand and specialize their smooth ER for Ca2+ storage and proliferate their mitochondria for energy production.
What defines the function of organelles?
Membrane lipid composition and proteins
Where are most proteins synthesised?
In the cytosol
What happens to proteins?
Need to move from ribosome to target site
Ribosome are attached to the cytosolic face of ER
Enter endomembrane system at ER
If membrane proteins will be integrated into membrane at this stage
If they are destined to be secreted or for lumen of another organelle – will be translocated across membrane
What do proteins destined for organelles have?
Sorting signals
What happens if a protein is destined to function in the cytosol?
It will just be synthesised and have no targeting sequence
What are sorting signals?
Short motifs with distinct amino acid sequences
Often found at the N terminus (usually removed from the finished protein by signal peptidases once sorting is complete)
ER - What are the 2 types of signal sequence?
a linear sequence of amino acids (called a signal sequence) or a specific three-dimensional arrangement of amino acids (called a signal patch)
Sorting signals for protein translocation into organelles are linear signal sequences, while examples of linear signals and signal patches are known for nuclear and vesicular transport
What are sorting signals for import into the nucleus?
Predominantly positively charged basic amino acids eg lysine
What are sorting signals for import into the ER?
linear sequence of about 5–10 predominantly hydrophobic amino acid (leucine, valines, isoleucine, phenylalanine, tryptophan)
What are sorting signals for proteins staying in the ER?
specific signal sequence of four amino acids at their C-terminus
Lys, Asp, Glu, Leu
What is the sorting signal for proteins destined for mitochondria?
positively charged amino acids alternate with hydrophobic ones
What % of all proteins in the cell are targeted to ER?
30%
What proteins are initially assembled at the ER?
Most soluble and integral membrane proteins destined for the cell exterior or for other organelles are initially assembled at the ER
o Soluble proteins are transported across the membrane
o Membrane proteins are inserted into the membrane
What are the pathways of the endomembrane system?
Secretory and endocytic
What other transport is via the ER?
Transport of some proteins to peroxisomes and lipid droplets
How is the endomembrane system organised?
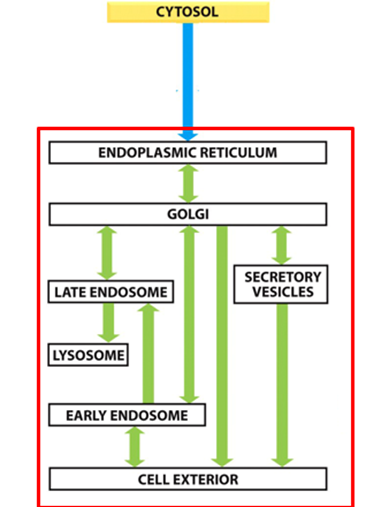
What is the secretory pathway?
o ER to golgi, sorted at trans-golgi
o Constitutive secretion = straight out
o Some will go into secretory vesicles where they will be stored until cells receive a signal
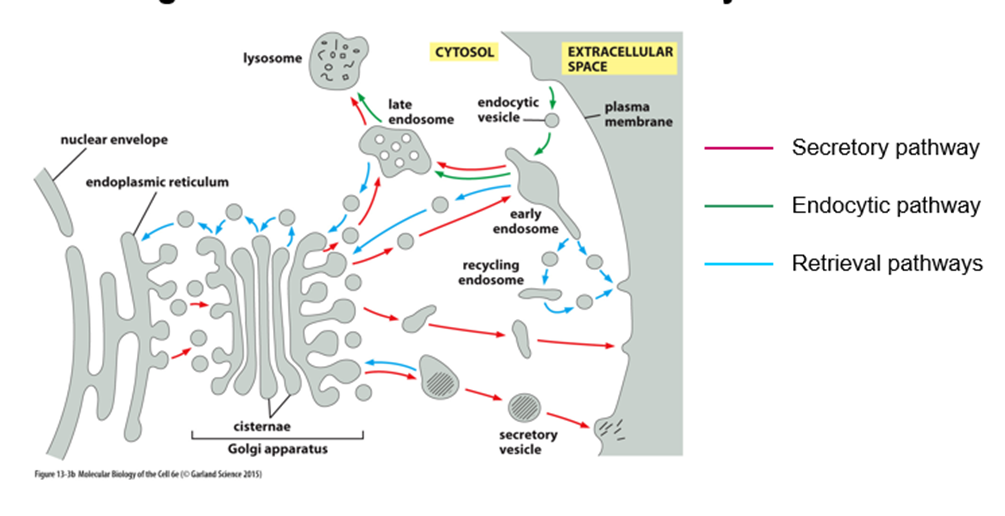
What is the endocytic pathway?
Opposite of secretory
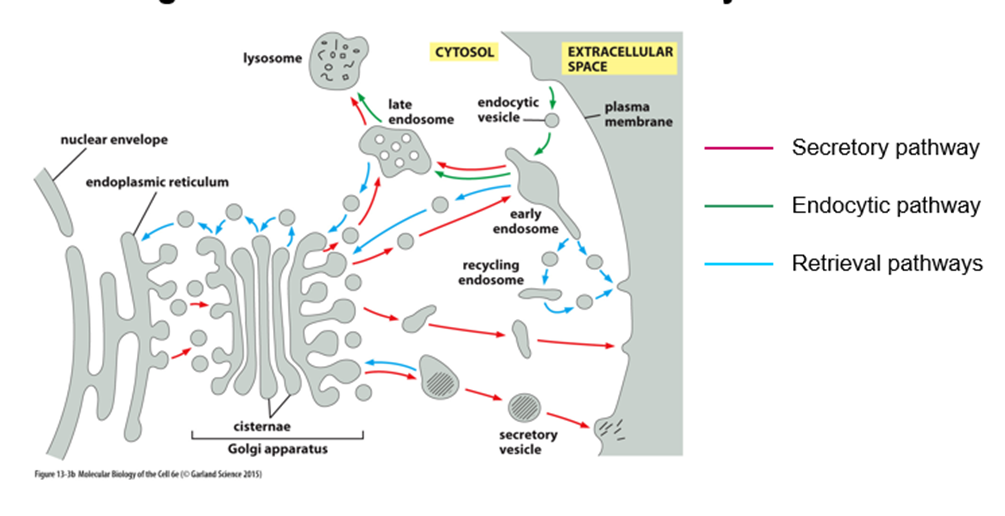
How is flow of membranes between compartments balanced?
By the retrieval pathway to maintain the membrane at a homeostatic level
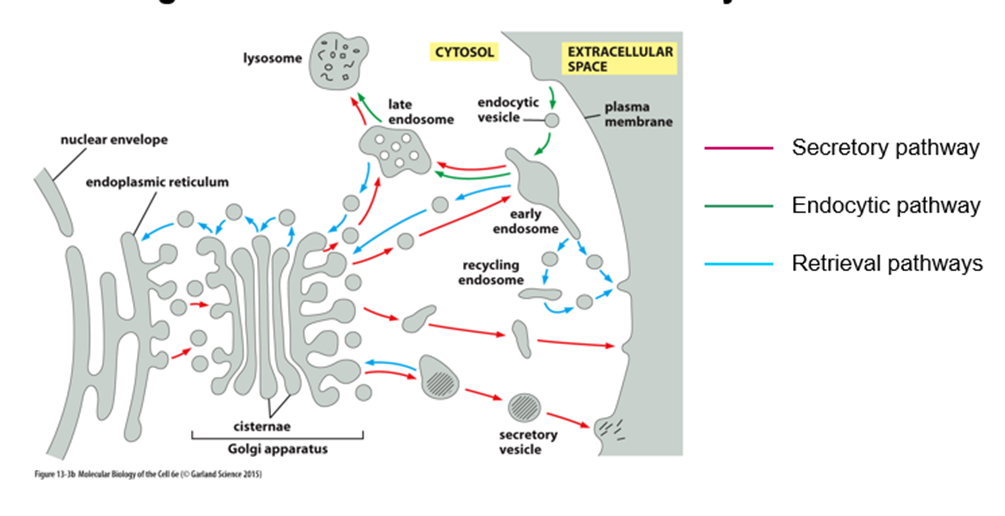
How do compartments retain distinct identity despite extensive flow of membranes?
They retain their lipid and protein composition
How is there transport between compartments?
It is discontinuous and mostly uses vesicles
Vesicles bud from donor compartment and fuse with acceptor compartment
Moves along cytoskeleton eg microtubules towards target compartment
Specific recognition event so vesicle will attach to surface of target compartment
Highly specific
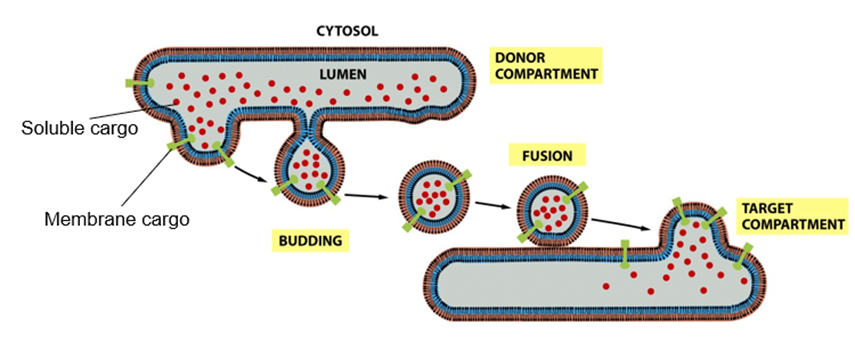
What is used if vesicular carries are not?
Tubular carriers
Still membrane enclosed by have different morphology
What is the same between vesicular and tubular carriers?
Discontinuous
Cargo is sorted into carrier
Machinery for movement - cytoskeleton
Machinery for fusion
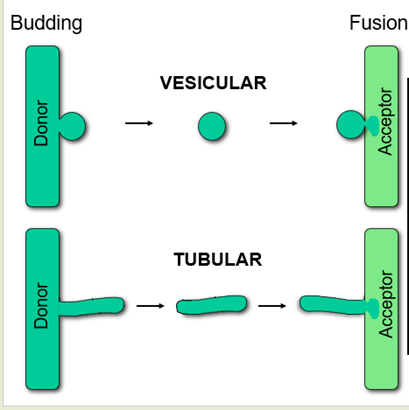
What is different between vesicular and tubular carriers?
Size and shape
Machinery for formation
Mechanisms of cargo sorting
True or false - Like the lumen of the endoplasmic reticulum, the interior of the nucleus is topologically equivalent to the outside of the cell
FALSE
• The nuclear interior and cytosol communicate through the nuclear pore complex and are technically continuous – not separated by a lipid bilayer
• In contrast, the lumen of the ER and the outside of the cell are separated from the cytosol by a lipid bilayer
Is it really true that ALL human cells have the same basic set of membrane-enclosed organelles? Do you know of any examples of human cells that do not have a complete set of organelles?
FALSE
The vast majority of cells in the human body do have the same basic organelles
But some cells lack some or even all organelles (RBC)
During its differentiation, the RBC precursor jettisons its membrane enclosed organelles, leaving only the plasma membrane and enclosed cytosol
Similar, is true for the lens of the eye, which lacks mitochondria
Some cells contain specialised organelles that are only found in that cell type or in cells that have a related function to that cell
Eg lysosome related organelles
What are the different approaches to identify transport machinery?
Biochemical approach
Genetic approach
Who suggested the biochemical approach?
Pioneered in the early 1980s by James Rothman for Golgi trafficking
Who suggested the genetic approach?
Pioneered in the early 1980s by Randy Schekman for the secretory pathway
What did James Rothman do?
Measured intra golgi transport in vitro using a cell free assay
Describe Rothman’s cell free assay
Took a cell in the lab which was mutant
Deficient in enzyme which adds sugar onto proteins
Infected with virus which makes glycoprotein which is transported through golgi (tritiated GlcNAc)
If he purifies Golgi membranes from these cells, they'll have the viral glycoprotein in the membrane But it won't have the sugar attached, because they lack the enzyme
Took 2 purified membrane fractions in a test tube (one had enzyme but not virus)
If viral glycoproteins goes over with no sugar added, it will be modified
Can measure the modification by using a radioactive sugar
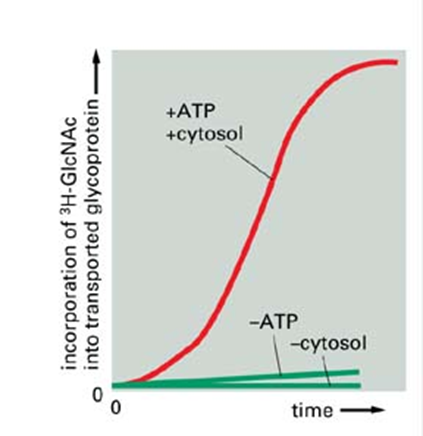
Transport was occurring, but it needed the presence of ATP
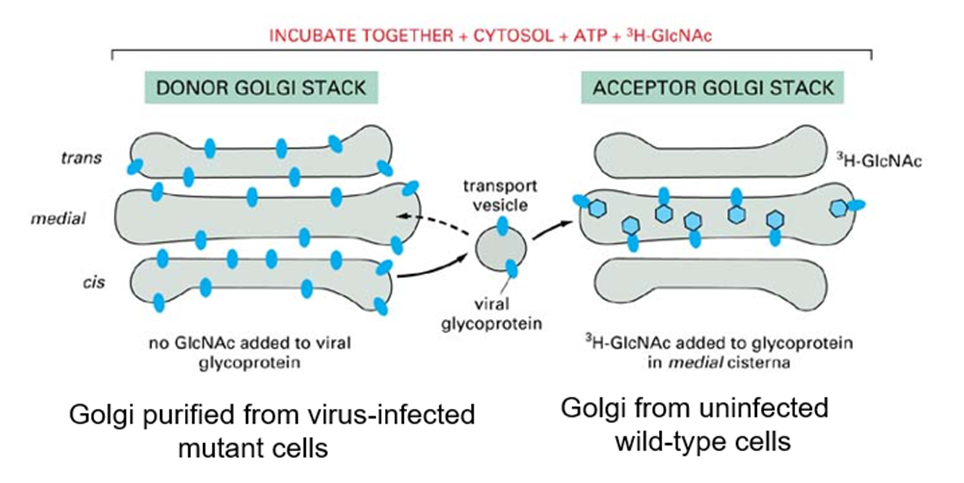
How did Rotham measure transport between golgi?
o Modification of viral glycoprotein
o Count radioactivity to measure transport
What did Rothman find transport of viral glycoprotein depended on?
Cytosol and ATP
What has Rothmans assay been used to identify?
identify a number of proteins required for budding and fusion of transport vesicles.
Important proteins were identified by fractionation of the cytosol
ie biochemical purification of transport factors based on their activity in the assay
What are the different types of cell-free systems for studying vesicular transport?
Permeabilise cells using a digitonin
Purified membrane fractions and cytosol
Purified proteins and lipids
What do cell free systems allow?
Allow manipulation of conditions to identify important parameters (e.g. requirement for temperature, energy, cytosolic and membrane proteins)
Have proved extremely useful for the identification of the machinery and mechanisms of transport
How did Schekman use yeast to identify mutants in vesicle transport?
Mutagenize yeast (eg with radiation)
This introduced lots of mutations into the genome
Look for mutants defective in transport
Got heavier as build up of proteins (couldn’t secrete them)
Density gradient
Why are yeast good model systems?
o They are easy to grow and divide rapidly
o Cheap to maintain
o They are amenable to genetic analysis (exist as haploids so if mutagenise gene will lose gene function)
How many different mutants did Schekman identify?
30
What did Schekman find about the mutants?
Mutants were temperature sensitive:
Function normally at permissive temperature (25˚C)
Defective in transport at non-permissive temperature (37˚C)
Because blocking secretion is lethal. Mutant yeast cannot grow under normal conditions
What were the mutants named?
Sec mutants
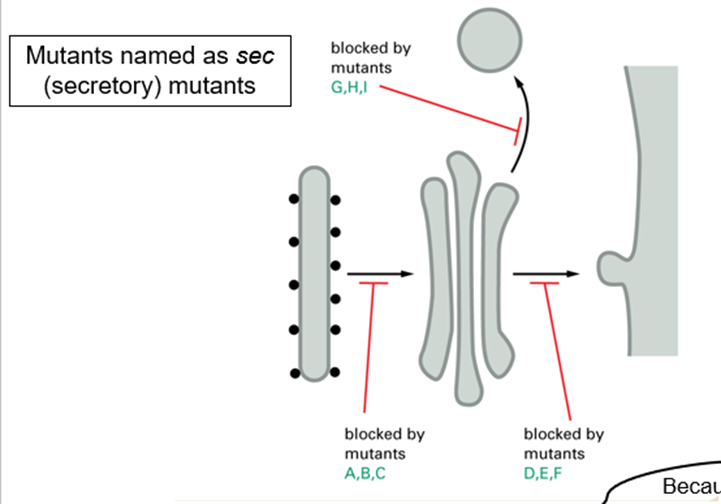
How do you determine whether a sec mutant is defective in budding or fusion?
Study distribution of cytoplasmic membranes
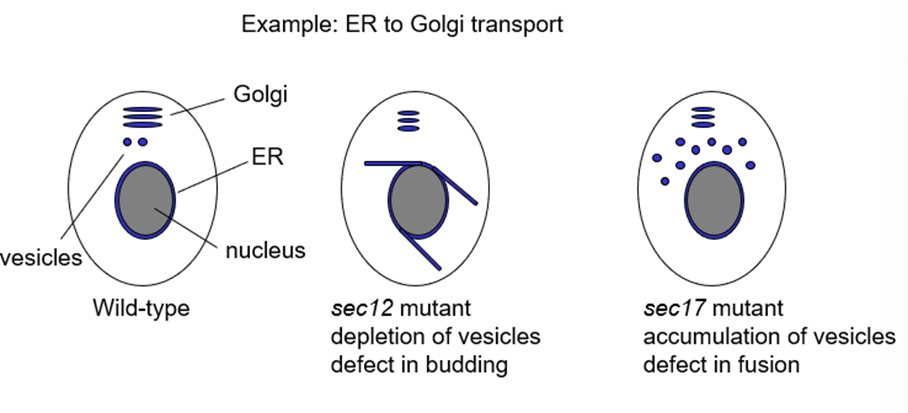
What does sec12 mutant do?
Required to make vesicles at ER so mutant shows depletion of vesicles
Defect in budding
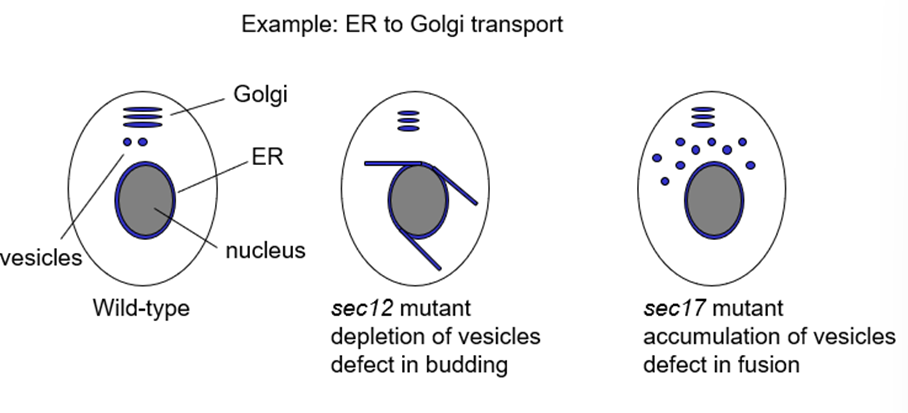
What does sec17 mutant do?
Required for membrane fusion of vesicles so sec17 mutant accumulates vesicles
Defect in fusion
A detailed description of the multicopy suppression method to identify mutant genes in yeast, as used in the identification of the Sec genes.
The yeast mutant e.g. Sec23, fails to grow at the non-permissive temperature because it has a secretion defect. There is a mutation in the Sec 23 gene that results in the production of a Sec23 protein that is non-functional at the non-permissive temperature because it cannot fold correctly at this temperature.
2. To restore growth, the yeast needs a functional wild-type Sec23 protein. However, when the experiment was done, the identity of this protein was not known.
3. So, to provide the yeast with Sec23, the mutant was transformed with the entire library of yeast genes. The library is comprised of thousands of DNA plasmids, each one containing roughly one yeast gene. Out of these thousands of plasmids, only a handful will contain Sec23.
4. The yeast transformants i.e. the mutant strain (Sec23) that has been transformed with the library, will be plated onto medium at the non-permissive temperature. They are plated at low density so that each yeast transformed with a single type of plasmid can grow into a colony. Only those yeast transformed with a plasmid encoding Sec23 will grow because only these will contain a functional Sec23 protein. Out of thousands of transformants, only a handful of yeast colonies will be present.
5. What is done now is that the plasmid DNA (containing the Sec23 gene) is isolated from each growing colony by standard methods and the DNA is sequenced. The sequence will correspond to the Sec23 gene, which encodes the Sec23 protein.
6. The same method would apply to all Sec mutants. Note that it is called multicopy suppression because multiple copies of each plasmid will exist within the yeast (because the plasmid replicates inside the yeast), and there is suppression of the mutant phenotype i.e. the mutant phenotype is suppressed (or you can think of it as rescued).
How do you identify the genes?
Multicopy suppression
Plasmid library of random normal genes transformed into mutant yeast strain
Grow yeast at non-permissive temperature
Yeast that survive have functional secretion - they express a gene that suppresses the defect
Isolate plasmid and sequence DNA to identify gene
How has GFP technology revolutionised cell biology?
• Naturally fluorescent protein (from jellyfish)
• Can be attached to any protein using DNA technology
• Allows the protein to be followed using live cell microscopy
• Barrel shaped with an active chromaphore in the middle
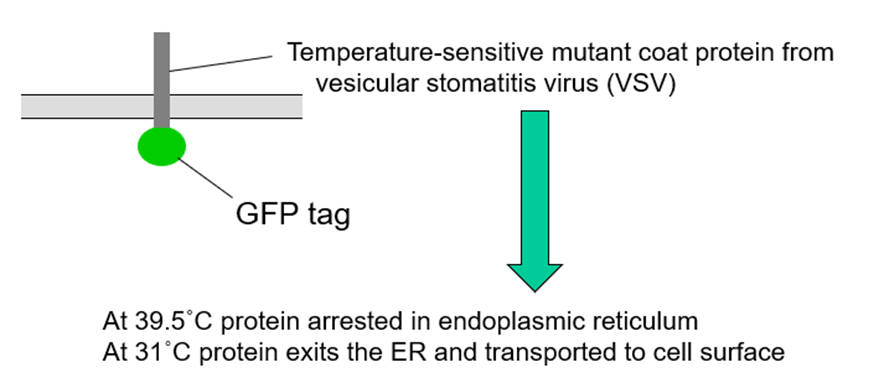
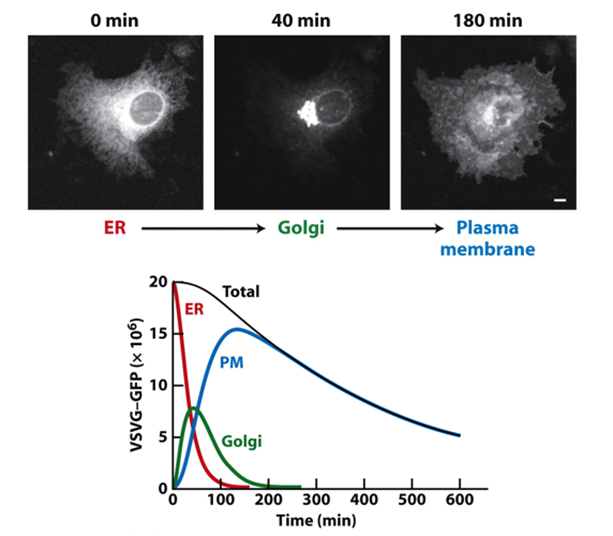
How does GFP show ER to golgi transport?
Experiment- shift cells VSV-GFP-expressing cells from 39.5˚C to 31˚C and analyse movement of the fluorescent protein using live-cell microscopy
Temperature sensitive mutant
AT 39.5 was stuck in ER as misfolded
If temperature dropped can fold
What is Super resolution microscopy?
Standard light microscopy resolution has a limited resolution (~200 nm), which is greater than the size of many cellular structures
Super-resolution microscopy uses various tricks to get round this issue resulting in resolutions down to ~20 nm i.e. 10x better
Types are SIM (structured illumination), STED (stimulated emission depletion), PALM (photoactivated localisation) and STORM (stochastic optical reconstruction) microscopy
What is electron microscope tomography?
• Allows 3D reconstruction of cellular organelles
• Tilt the sample to build up view
• Uses thick sections
1.) In the Rothman Golgi transport assay, the investigators hypothesised that the viral glycoprotein was transported between the Golgi stacks in vesicles. An alternative hypothesis is that the viral glycoprotein was released from one Golgi apparatus and taken up by the other, without being packaged into vesicles. Which of the following experiments would best distinguish between these 2 hypotheses:
A. Determine whether transport still occurs without energy (ATP).
B. Test for association of the modified viral glycoprotein with membranes by centrifugation.
C. Test whether transport between the Golgi stacks is blocked by addition of a detergent.
D. Add a protease to the system and determine whether the viral glycoprotein is degraded.
D
The Schekman assay for identifying trafficking machinery used the technique of multicopy suppression to identify the SEC mutant genes. This method is ‘old’ technology. Can you think of how we might identify the mutated genes nowadays using current genomic technologies?
The current standard method would be to isolate the genomic DNA of the cells of interest eg SEC mutant cells, and perform DNA sequencing to sequence the entire genome. This is fast and cost effective and allows direct identification of any mutations and which genes they are found in.
Pros of GFP
o Can directly visualise the protein of interest – no need for antibody
o Can be used in live cells so can visualise dynamics and avoid fixation artefacts
o Technically easy to tag protein of interest and image it
Cons of GFP
o Large – 27kDa so could interfere with folding or function of protein
o Could interfere with assembly of multisubunit protein complexes
o Need to express the protein so if over expressed could cause localisation artefacts