Enzymes and coenzymes
1/14
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
15 Terms
Enzymes basic facts
Biological catalysts
increase the rate of reaction by lowering activation energy
Nearly all proteins, some are RNA (riboenzymes)
Active site = area of specific AA residues
Many have -are suffix
7 major classes of enzyme
Oxidoreductase
transfer of electrons in redox reactions E.g. dehydrogenases
Transferase
transfer function groups E.g. RNA polymerase
Hydrolase
hydrolysis - water added E.g. proteases, ATPases
Lyase (Synthase)
group removal, cleave bonds by elimination to form new products E.g. decarboxylase
Isomerase
rearrangement of atoms in molecules E.g. cis-trans isomerases
Ligase (Synthetases)
joining two molecules, needs energy E.g. DNA Ligase
Translocases
movement of ions or molecules across membranes E.g. pumps
Examples of industrial/clinical uses
Subtilisin = extracellular serine endopeptidase, biological action of washing powders
Taq DNA Polymerase = used in PCR for amplification of DNA
Calf rennet = mix of proteases, separates milk curds and whey
Liver function tests = Alanine aminotransferase (ALT)
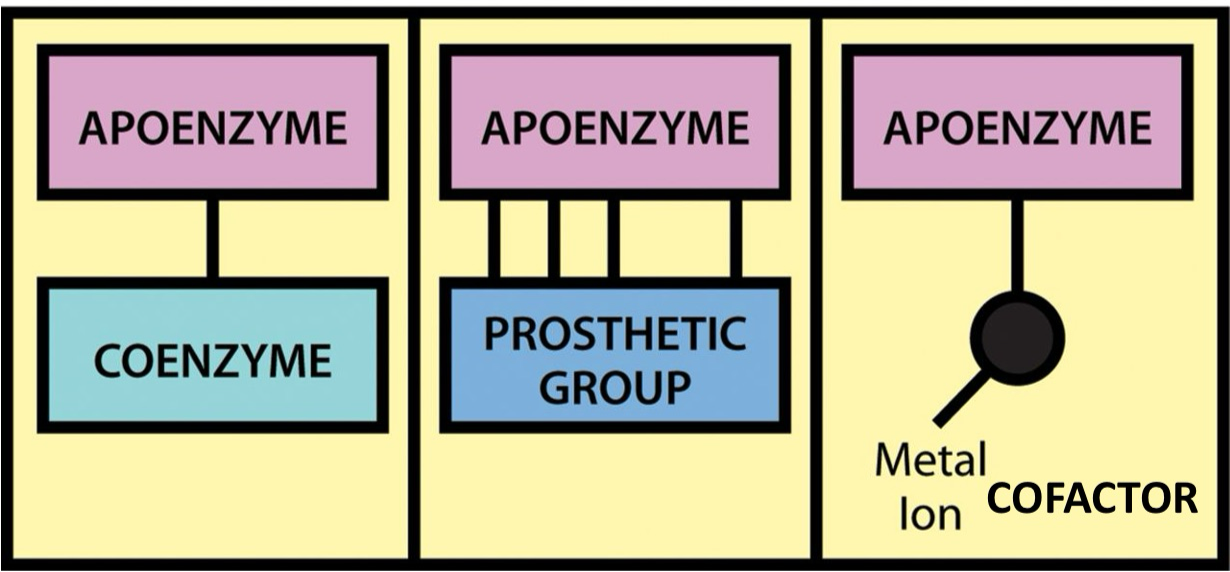
Coenzymes and Cofactors
small non-protein molecule required for enzyme fun
Cofactors = inorganic ions e.g. Fe2+
Coenzymes = organic molecule e.g. ATP, FAD
Cosubstrates = loosely bound type of coenzyme, bind and unbind during catalysis
Prosthetic group = covalently bound cofactors/enzyme
Holoenzyme = catalytically active with its cofactor/enzyme
Apoenzyme = inactive, only protein
Basic thermodynamics
Cells, tissues and organs = systems
Entropy (S) = degree of randomness/disorder
2nd law of thermodynamics = change in entropy of system + change of entropy in surroundings = 0
Difficult to measure in biochemical processes
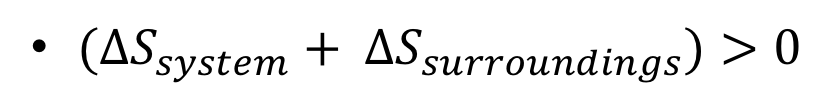
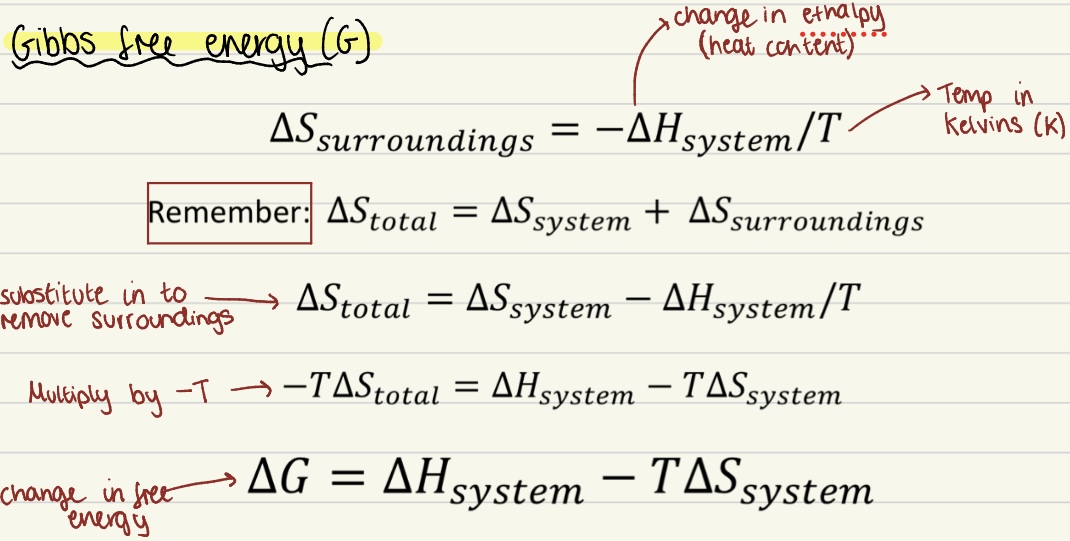
Gibbs Free Energy (G)
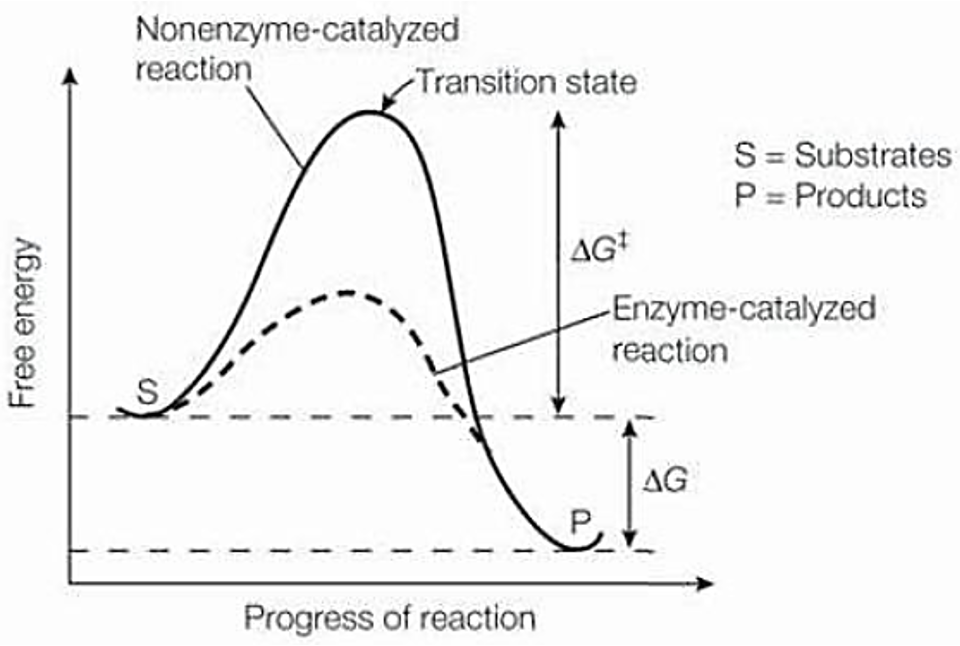
How do enzymes lower activation energy
Enzymes lower the activation energy (change in GFE)
Enzymes bind tightly to X (transitions state) not S or P
S→X→P
GFE of activation
A measure of the energy barrier that must be overcome for the reaction to proceed at a certain rate
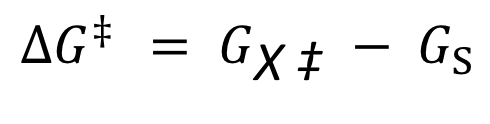
Components of a holoenzyme
Enzyme catalytic strategies
Covalent catalysis
Active site contains reactive groups, which covalently bonds to the substrate during the reaction
General acid-based catalysis
transfer of protons (donate or accept) to or from and intermediate
Catalysis by approximation
binding surface of enzyme brings two substrates into close proximity so a reaction can happen
Metal ion catalysis
ions can act as an electrophile (electron acceptor) and stabilise negative charge
Facilitate nucleophile formation by coordination
Act as a bridge between enzyme and substrate
Proenzymes/Zymogens
Enzymes synthesised as larger inactive precursors
Zymogens activated by proteolytic (hydrolytic) removal of a peptide
E.g. trypsin ‘cuts’ lots of enzymes to activate them
Require a biochemical change to activate
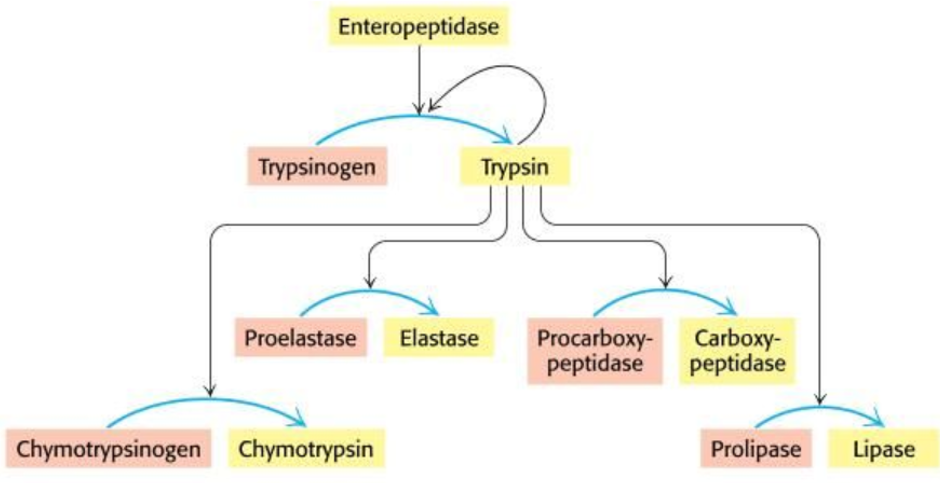
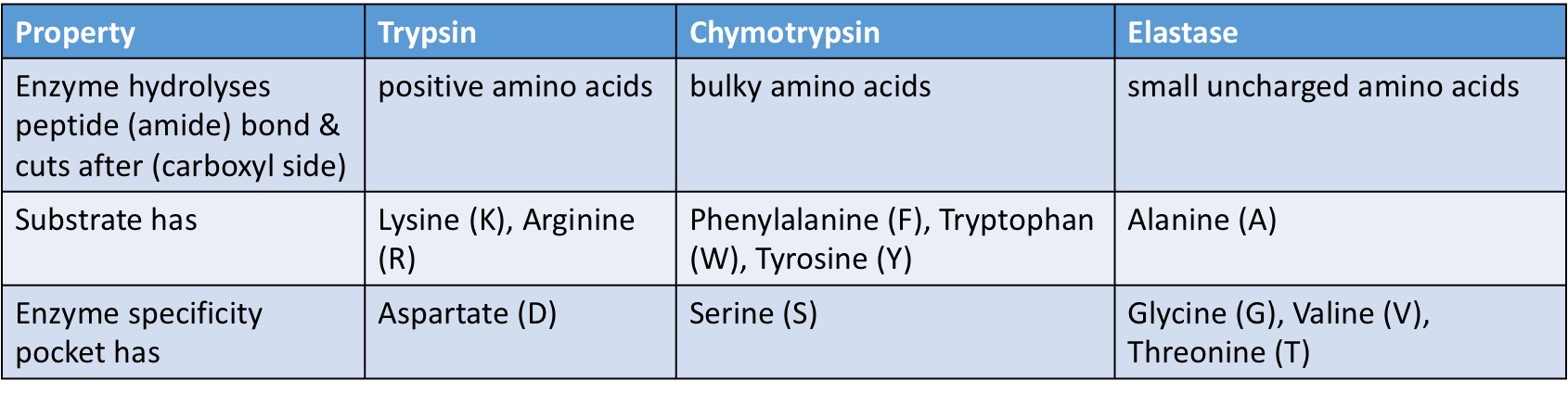
Substrate specificity of endopeptidases
Endopeptidases → split peptide bonds in proteins creating smaller proteins
Catalytic triad of Chymotripsin
Covalent catalysis→ impermanent covalent bond between enzyme and substrate
Histidine 57 is a proton acceptor (nucleophile)
Pulls H+ from O-H bond of Serine 195
Carboxyl group of Aspartate hydrogen bonds with histidine to orient it to be a proton acceptor
Two phase process
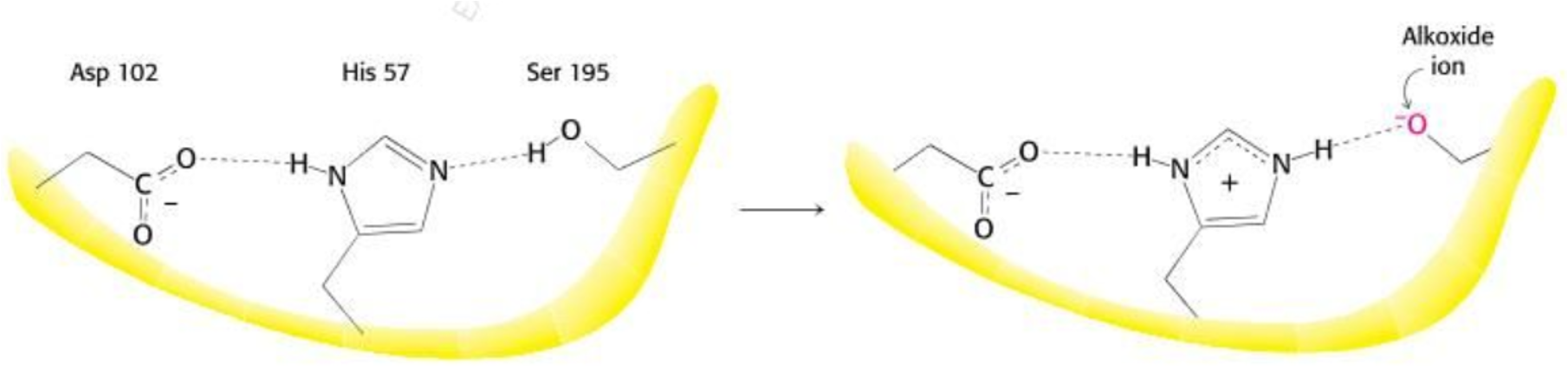
Chymotripsin → Acetylation
Aspartate 102 helps orient Histidine, which then acts as a base to pull a H from Serine’s alcohol group
Activated Serine attacks the carbonyl carbon of the peptide bonds on the substrate
This forms a temporary, unstable ‘tetrahedral intermediate’
Intermediate collapses, Histidine donates a H+ which breaks the peptide bond and releases the C-terminal portion of the peptide
The N-terminal portion stays covalently bonded to Serine
Chymotripsin → Deacylation
A water molecule enters the active site
Histidine removes a H from the water molecule, making it a strong nucleophile
Activated water molecule attacks the carbonyl carbon on the Acyl-enzyme
A second intermediate collapses, Histidine donates its proton back to Serine, breaks bond between enzyme and substrate
N terminal released and active site regenerates