BIOB10 - Cell Biology - Midterm
5.0(1)
Card Sorting
1/114
There's no tags or description
Looks like no tags are added yet.
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
115 Terms
1
New cards
DNA
Stored information in double helix
Nucleotide: sugar phosphate + base (a,c,t,g)
Sugar phosphate backbone, bases connect but aren’t stuck together
Nucleotide: sugar phosphate + base (a,c,t,g)
Sugar phosphate backbone, bases connect but aren’t stuck together
2
New cards
DNA replication
Complimentary strand, templated polymerization, hydrolysis aids, monophosphate attaches.
3
New cards
Templated polymerization
Using another strand to make another strand. Using a template to make a complimentary strand.
4
New cards
Hydrolysis
GTP → High E bonds break → GDP + energy
Addition of water
Addition of water
5
New cards
Nucleotide
Nucleoside with extra one or more phosphate group attached (monoguanosine).
6
New cards
Nucleoside
The sugar and the backbone with phosphate group mentioned
7
New cards
Transcription
Cell needs to make sense of information. Take one strand and use it as a template to form RNA strand.
DNA → RNA
Read one at a time, RNA called transcript. Complimentary base pairings (T = U)
DNA → RNA
Read one at a time, RNA called transcript. Complimentary base pairings (T = U)
8
New cards
Translation
“What amino acid do I add?”
RNA → Protein
Read three nucleotids (1 codon) at a time. 1 codon = 1 amino acid. Ribosome does readings.
RNA → Protein
Read three nucleotids (1 codon) at a time. 1 codon = 1 amino acid. Ribosome does readings.
9
New cards
Plasma membrane
Cell is closed in. Selective barrier → how things move in and out, semi-permeable
Amphiphilic → hydrophobic tails face inward, hydrophilic head towards water
Amphiphilic → hydrophobic tails face inward, hydrophilic head towards water
10
New cards
Eukaryotes
House genetic information in nucleus. Bigger genome, difference in cell structure.
11
New cards
Macromolecules
Like protein or nucleic acid.
Fold into energetically favourable confirmation, what makes the most sense.
Fold into energetically favourable confirmation, what makes the most sense.
12
New cards
Enzymes
Proteins that facilitate certain reactions. Like DNA polymerase (hydrolysis happens and it takes nucleotide away)
13
New cards
Nuclease
Enzyme that cuts nucleic acid
Exo: cutting from end
Endo: snipping from middle
Exo: cutting from end
Endo: snipping from middle
14
New cards
Polypeptide chain
Multiple amino acids connected by peptide bonds.
Backbone (peptide bonds connecting), Side chain.
N-terminus front end (amino group), C-terminus back end (carboxyl group)
Backbone (peptide bonds connecting), Side chain.
N-terminus front end (amino group), C-terminus back end (carboxyl group)
15
New cards
Hydrophobic clustering
Dropping protein into an aqueous watery environment causes proteins to move to protect parts from water.
Non-polar parts are hydrophobic.
Non-polar parts are hydrophobic.
16
New cards
Protein denaturing
Treat proteins with a solvent to mess up their bonds (unfold)
17
New cards
Protein renaturing
Remove solvent and protein folds again
18
New cards
Molecular chaperones
Proteins that help proteins fold properly. Folding helpers.
They do not determine shape, just help form it. Side chain interactions determine shape, exposed hydrophobic parts are meant to fold inward.
They do not determine shape, just help form it. Side chain interactions determine shape, exposed hydrophobic parts are meant to fold inward.
19
New cards
Off-pathway conformations
Exposed hydrophobic regions can result in this. Desperately going around and binding to stuff and making a mess.
20
New cards
Protein structures
Coiled-coil (hydrophobic face eachother), Protein complex (protein subunits join together, clump), Dimer (2, head to head arrangement. chains bind together)
21
New cards
Protein characteristics
Fibrous (long, coiled-coil, helix) OR globular (clump, DNA polymerase)
Can be disordered in regions (nuclear pores, gates)
Tight or weak
Specificity
Can be disordered in regions (nuclear pores, gates)
Tight or weak
Specificity
22
New cards
Ligand binding
Referring to something that is doing bonding. Like enzymes, often first step of what an enzyme does. Fits with binding site.
23
New cards
Ligand
Surface contours of the molecule fit very closely to the protein. Sending communication from one molecule to another (like to change formation)
24
New cards
Protein binding interfaces
Surface-string, helix-helix, surface-surface (most common)
25
New cards
Gated transport
Pores where things can go in and out. Referring to nuclear pore complex.
26
New cards
Protein translocation
Proteins need to get to a destination; transmembrane proteins.
The protein uses transmembrane protein to snake through.
The protein uses transmembrane protein to snake through.
27
New cards
Vesicular transport
(main mean of protein transportation) Take a vesicle that transport from one place to another.
28
New cards
Signal sequences
Sorting signal. Sequences on the N-terminus or in the middle of the polypeptide chain.
29
New cards
Cytosolic protein
No signal sequence, is just in the cytoplasm
30
New cards
Co-translational translocation
Cells move proteins into the ER before completion of polypeptide synthesis.
ER sequence shows up and protein synthesis continues on ER as chain gets snaked into ER
ER sequence shows up and protein synthesis continues on ER as chain gets snaked into ER
31
New cards
Post-translational translocation
Finishes making protein and THEN goes to where it needs to be.
32
New cards
ER signal sequence
Guided to the ER by two main parts.
Signal-recognition particle (SRP) and SRP receptor.
Signal specifically on N-terminus.
Signal-recognition particle (SRP) and SRP receptor.
Signal specifically on N-terminus.
33
New cards
Signal recognition process
SRP binds to ER signal sequence, SRP bends and block elongation factor site (pauses translation). SRP receptor binds SRP and it moves whole junction of ribosome and SRP down to ER. SRP receptor bonding. Protein elongates through protein translocator while ribosome is released.
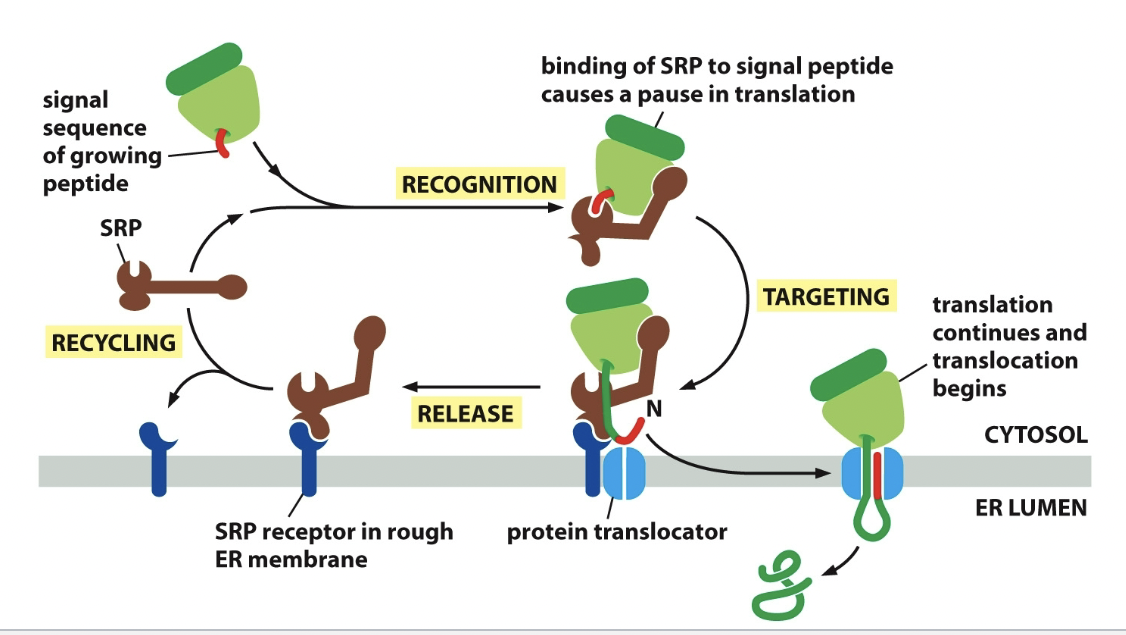
34
New cards
Secretory pathway
Where stuff is going to get closer and closer to exterior of the cell
35
New cards
Membrane bound ribosomes
Physically on the ER, doing translation on the ER.
36
New cards
Free polyribosomes
Transcript with 5’ and 3’ end (read from 5’)
Ribosome keeps reading and moving down. Second and third ribosomes don’t wait till first one is done, they’ll go when there’s enough room.
Once done, ribosome disassembles into individual subunits.
Ribosome keeps reading and moving down. Second and third ribosomes don’t wait till first one is done, they’ll go when there’s enough room.
Once done, ribosome disassembles into individual subunits.
37
New cards
Membrane-bound polyribosomes
ER signal sequence, SRP bounding to ER membrane. Continues elongating as protein snakes through. As soon as there is space, the next one attaches and binds to membrane next to first ribosome.
38
New cards
Sec61 complex
Core of translocator. Has a plug, when it recognizes the sequence, the plug moves aside and allows protein to enter.
Signal peptide is hydrophobic and cannot snake through
Signal peptide is hydrophobic and cannot snake through
39
New cards
Accessory proteins
Work with translocator to help with the process of protein translocation.
BiP
BiP
40
New cards
BiP
Binding protein in the ER. ATP → ADP through hydrolysis. Happens in order for this to pull the protein through.
41
New cards
Translocon
Translocator + accessory proteins.
42
New cards
Targeting
(part of SRP cycle) Start-transfer sequence, signal sequence doesn’t need to stay and can be cut off.
Stop transfer.
Stop transfer.
43
New cards
Start-transfer sequence
It is recognized by ribosome, it opens up the translocator and enters.
44
New cards
Stop transfer
Get out and the little protein is embedded in the membrane
45
New cards
Single-pass transmembrane protein
Goes into membrane, doesn’t get cleave off, serves as an anchor. With or without stop transfer sequence. It is stuck in membrane.
46
New cards
Multi-pass transmembrane protein
Rest of protein snakes in, no stop transfer, just goes straight through and it’s done. Double pass if there is a stop transfer.
47
New cards
ER lumen protein
If they wanna be right inside the ER. One signal sequence, cleaved off by signal peptidase. Snakes right through.
48
New cards
ER resident proteins
Stays in the ER for whatever reason. BiP is one. Have and ER retention signal.
49
New cards
Protein quality control
ER must make sure protein folds correctly.
Done with Oligosaccharides and ER chaperones.
Done with Oligosaccharides and ER chaperones.
50
New cards
Oligosaccharides
Added to proteins. Adds three sugars to proteins (protein glycosylation), catalyzes (transferase).
51
New cards
ER chaperones
Proteins help proteins fold properly.
Lectins (calnexin and calreticulum)
Bind to oligosaccharides on incompletely folded proteins and retain them in ER.
Lectins (calnexin and calreticulum)
Bind to oligosaccharides on incompletely folded proteins and retain them in ER.
52
New cards
Oligosaccharides processing
Adding 3 glucose to unfold protein. Glucosidase trim these off one at a time.
53
New cards
Retrotranslocation
The cutting process. Protein folding process doesn’t work out, it goes around the second time.
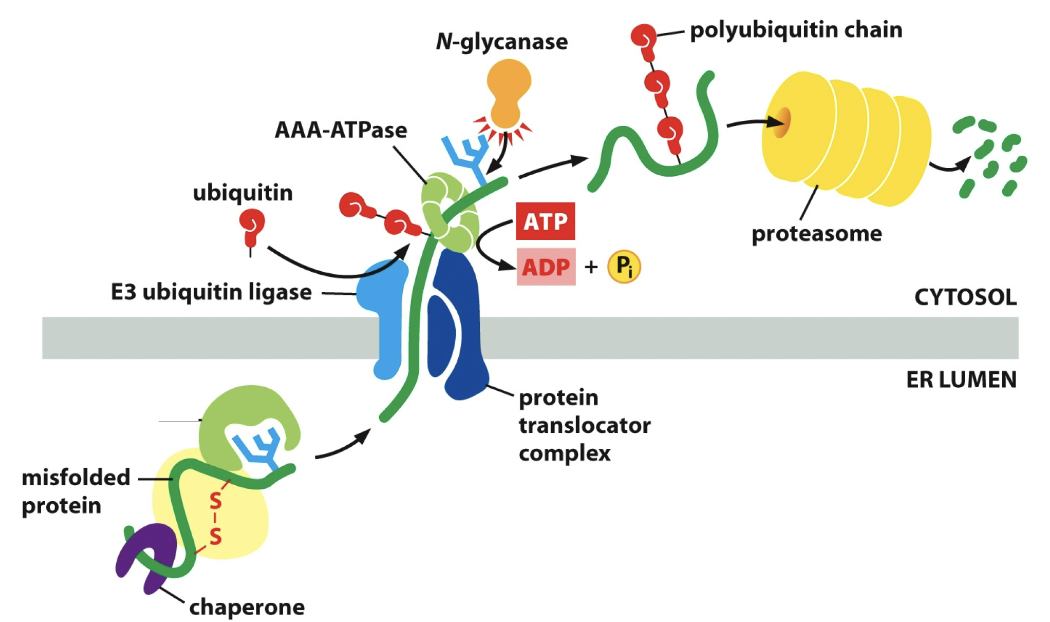
54
New cards
Mannosidase
Removes mannose on the core oligosaccharide tree. Not gonna get glucose added back. Mannose is removed. Protein without mannose gets destroyed.
55
New cards
E3 ubiquitin
enzyme. Stick onto the protein and protein translocator complex. Stamps it to be destroyed.
56
New cards
AAA-ATPase
Like BiP, helps pull protein out. Does this by hydrolyzing ATP. In retrotranslocation.
57
New cards
N-glycanase
Enzyme, still has the ubiquitin chain. It saves the chain. In retrotranslocation.
58
New cards
Heat-shock response
Wants more chaperones to be made, needs to go all the way back to the DNA to get that coding sequence.
Misfolding with a lot of heat → stimulates transcription of genes encoding cytosolic chaperones that help refold proteins
Misfolding with a lot of heat → stimulates transcription of genes encoding cytosolic chaperones that help refold proteins
59
New cards
Unfolded protein response
Making more proteins involved in retrotranslocation, protein degredation in the cytosol, and ER chaperones
60
New cards
Protein translocators
Doors/gates that allow protein to pass.
Mitochondria → TOM, TIM(22/23), SAM, OXA complexes
Mitochondria → TOM, TIM(22/23), SAM, OXA complexes
61
New cards
TOM complex
Transfer proteins across the outer mitochondrial membrane. Initial recognition.
62
New cards
TIM complex
Transfer proteins across inner mitochondrial membrane. Continue journey down, helps bring the protein down.
63
New cards
SAM complex
Once protein enters TOM, this helps them fold properly in beta porin
64
New cards
OXA complex
Some proteins can be made in mitochondria and it does that.
Helps protein snake through, get’s cleaved in matrix. This will help it and stick it in inner membrane.
Helps protein snake through, get’s cleaved in matrix. This will help it and stick it in inner membrane.
65
New cards
Cytosolic hsp70
(importing to mitochondrial matrix) Interacting with protein, gonna keep it unfolded. Helps that interaction. Comes off once recognized by translocator. Binds on and off to fish through.
66
New cards
Mitochondrial hsp60
Chaperone protein. Interacts with protein and once it comes through, this helps it fold through ATP hydrolysis.
67
New cards
Chloroplast
Post-translationally, translocation, ATP hydrolysis, uses TOC/TIC, added thylakoid.
68
New cards
Turnover number
Where the reaction rate will max out
69
New cards
Lysozyme
Enzyme that cut cell walls of bacteria, cell bursts. Needs to be in transition state, bound to enzyme.
70
New cards
Regulatory sites
Small molecules bind to these. Separate from active sites. Recognizes a regulatory molecule.
71
New cards
Active site
Part of enzyme that recognizes substrate, and then stuff changes when it binds with stuff.
72
New cards
Positive regulation
Inactive → active (both easily bind)
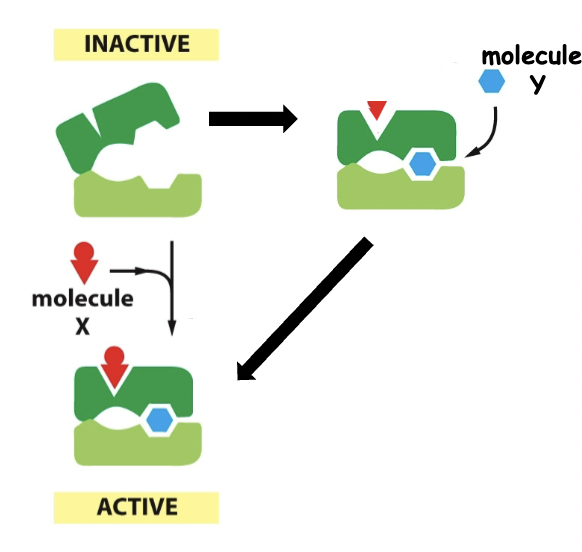
73
New cards
Negative regulation
Active → inactive (molecules don’t want the same thing)
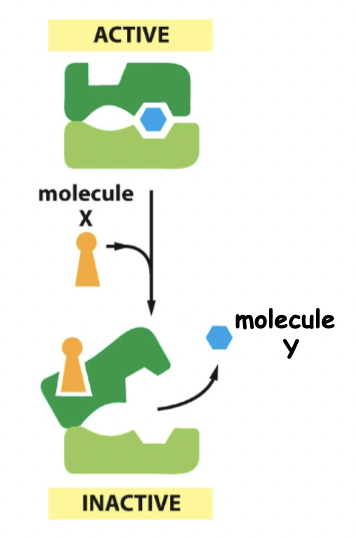
74
New cards
Cooperative allosteric transitions
Binding of one inhibitor at a time, opens it up one subunit at a time. First inhibitor binds with difficulty (changes confirmation of second inhibitor in the process) and second inhibitor binds with ease.
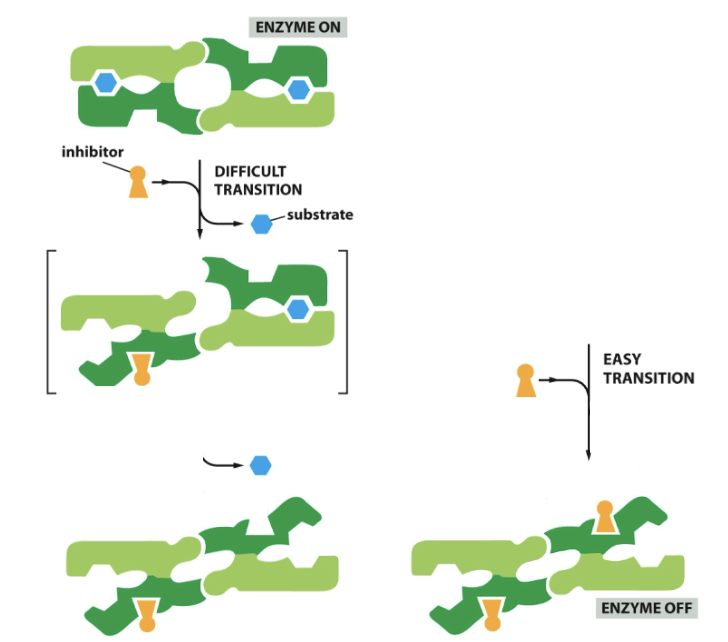
75
New cards
Endocytosis
Pinching in of plasma membrane. Removes plasma membrane components and delivers them to internal compartments called endosomes.
76
New cards
Endocytic pathway
Leads inward from plasma membrane
77
New cards
Exocytosis
Secretory pathway delivers new proteins to plasma membrane or extracellular space.
78
New cards
Secretory pathway
Leads outward from ER towards Golgi and cell surface (or side route to lysosomes)
79
New cards
Coated vesicles
Cage of protein covering cytosolic surface, discard cote before fusing. Coat has 2 layers.
80
New cards
Clathrin-coated vesicles
Golgi to plasma membrane, 3 heavy and 3 light chains form outerlayer (triskelyons, make buds), clathrin protein component, adaptor proteins (bind to coat and trap proteins), PIPs
81
New cards
PIPs
Bind to AP2s and change adaptor proteins conformation. Can become phosphorylated at different regions and then bind to APs and change them. Then it is possible to bind to receptors.
They control when protein is active or inactive (no coat when inactive)
They control when protein is active or inactive (no coat when inactive)
82
New cards
COPI
Coated transport vesicle, Golgi → ER
Arf involved
Arf involved
83
New cards
COPII
Coated transport vesicle, ER → Golgi
Sar1 involved
Sar1 involved
84
New cards
Membrane bending proteins
Have crescent shaped domains (BAR domains). Bind to and impose shape, help form circular bud.
85
New cards
Release from membrane
During uncoating of membrane, PIP that is packed into vesicles leave, this weakens adaptor protein’s bind. Adaptor proteins uncoat, as soon as they pinch off coat is useless. Uses uncoating ATPases
86
New cards
GEFs
GDP → GTP
Molecular switch, GTPases, activate protein, turn on
Molecular switch, GTPases, activate protein, turn on
87
New cards
GAPs
GTP → GDP
Molecular switch, GTPases, inactivate protein, turn off.
Molecular switch, GTPases, inactivate protein, turn off.
88
New cards
Sar1
Inactive bound to GDP, needs to work with molecular switch. GEF takes inactive protein and activates it.
89
New cards
Arf
Active, bound to GTP. Works with GAPs to inactivate.
90
New cards
Rab-proteins
GTPases, plays a role in vesicle arriving at correct target membrane.
__-GDP is inactive
__-GEF binds and activates protein (GDP → GTP)
Tail unfolds into membrane when activated (anchor)
__-GDP is inactive
__-GEF binds and activates protein (GDP → GTP)
Tail unfolds into membrane when activated (anchor)
91
New cards
Rab effectors
Rab-GTP works with them, they’re tethers, catch vesicles and bring em in. Tethering proteins. Long-threadlike.
92
New cards
SNARE proteins
Responsible for vesicles completing membrane fusion (lipid bilayers merging to dump contents)
V: found on vesicle with 1 protein
T: found on target membrane with 3 proteins
V: found on vesicle with 1 protein
T: found on target membrane with 3 proteins
93
New cards
trans SNARE complex
v- and t-SNARES wrap around to form a stable 4 helix bundle; locks the 2 membranes together for membrane fusion.
94
New cards
Vesicular tubular clusters
Two vesicles have their trans-SNARE complexes separated by NSF. Two vesicle’s clusters bind together and then have membrane fusion
95
New cards
Retrograde transport
COPII vesicles going from ER to Golgi, fusing into a large tubular cluster
Things need to go back to ER as cluster or individual (ER resident protein or COPI)
Things need to go back to ER as cluster or individual (ER resident protein or COPI)
96
New cards
Retrieval pathway
ER resident proteins have retrieval signals if they’re sent out. BiP has KDEL sequence on C-terminal signal. Retention mechanism (ER resident protein bind to each other and form complexes too big to enter).
97
New cards
Golgi pathway
CGN → cis cisterna → medial cisterna → trans cisterna → TGN → lysosomes
98
New cards
Secretion
Leaving TGN. Destined for lysosomes (digestive enzymes), exocytosis (sent to cell membrane, straight to cell surface)
99
New cards
Lysosomes
Membrane enclosed organelles filled with enzymes that digest macromolecules. There is proteases and nucleases within them.
100
New cards
Acid Hydrolases
Enzymes in lysosomes need an acidic environment to function. Controls them from going and destroying everything. They’re physically contained and the pH isn’t suitable for them.