DNA, RNA and gene expression
1/96
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
97 Terms
Purine
A type of nitrogenous base with a double-ring structure, which includes adenine (A) and guanine (G).
Pyrimidine
A type of nitrogenous base with a single-ring structure, which includes cytosine (C) and thymine (T).
Helicase
An enzyme that unwinds the double-helix structure of DNA during the replication process.
Ligase
An enzyme that joins the ends of DNA strands by forming new phosphate bonds.
Exonuclease
An enzyme that removes nucleotides from the ends of a DNA strand, functioning in both 3’ to 5’ and 5’ to 3’ directions.
Origin of replication
The specific location on the DNA molecule where replication begins.
In EUKARYOTES there are multiple origins replication.
Bidirectional in Eukaryotes
Bases added at a rate of 1000/second
So replication of the whole mammalian genome takes approximately 8 hours
In PROKARYOTES there is only ONE origin of replication
Telomere
The repetitive DNA sequences located at the ends of linear chromosomes, important for maintaining chromosome integrity.
G:C rich repeats
In humans - TTAGGG/AATCCC - highly conserved
NON-CODING but ESSENTIAL to maintain the integrity of DNA
Nucleoside
sugar + base
Phosphate attaches to 5' carbon
1st Carbon binds to base through N-glycosidic bond = stability to sugar and base (nucleoside)
Anti-parallel
Bonding between bases
Different H-bonds between bases allows a twist to the antiparallel structure = DNA double helix. Compacted in such a way, it can be unravelled for eukaryotic processes
Major/minor groove
Sequences that are accessible on outside of major groove allows transcription factors or replication machinery to actually bind into the minor or major groove
Phosphate on outside of two antiparallel strands and bases are in middle = helix can form
5’ to 3’
5' = phosphate group
3' = free hydroxyl group
Asexual reproduction - prokaryotes
The entire genome is on one circular chromosome = DNA molecule
The chromosome replicates once to produce two chromosomes that are identical (except for rare mutations).
The two identical daughter chromosomes move toward opposite end of the cell.
When the cell divides the daughter chromosomes are partitioned one to each daughter cell.
Asexual reproduction - eukaryotes
DNA replicates during S
Gene expression occurs during G1 and G2 (and S?)
Nuclear division (mitosis) occurs during Mitosis
Cell division (cytokinesis) occurs at the end of Mitosis
Some genes may be expressed in S phase but they will be genes that aren't highly expressed
Highly expressed genes will be replicated early in S phase - replication and transcription can't happen simultaneously
Silent genes are replicated late so there can be some transcription
General features of DNA replication
•semi conservative
•It is bidirectional process
•It proceed from a specific point called origin
•It proceed in 5’-3’ direction
•It occur with high degree of fidelity
•It is a multi-enzymatic process
occurs by three steps
1. Initiation
2. Elongation
3. Termination
Single-strand binding protein
binds to and stabilizes the single strand to keep DNA unwound.
bind on either strand
Primase
adds ribonucleoside triphosphates to synthesise an RNA primer
Binds at the initiation point of the 3'-5' parent chain
Bind to 3' to 5' parent strand because primase runs in 5' to 3' direction to lay down the primer
DNA polymerase
proceeds in a 5’ to 3’ direction
Adds 1000 bases /second to the growing chain
Requires all 4 dNTPs deoxyribonucleotides
MUST have a template and a primer
Has proof reading activity - Will shuffle back and check its laid down the right bases = fidelity
attach to RNA primer
Initiation
Primase lays down a primer
Single strand binding proteins are going to bind to each of the parent strands
In a 5' to 3' direction on both strands and therefore using 3' to 5' parental strand as template
elongation
DNA polymerase extends in a 5’ to 3’ direction
DNA polymerase attaches to RNA primer and then lays down comp. bases according to parent strand in 5' to 3' direction on both strands
Exonuclease removes the RNA primer
Polymerase fills the gap
Ligase joins the two pieces ia a condensation reaction forming 3' to 5' phosphodiester bonds
Can remove a primer because it is RNA and therefore has a different sugar and bases
Okazaki fragments
The lagging strand has to wait for the helicase to break the H-bonds before it can lay down an RNA primer for the DNA polymerase to replicate. Therefore it's made up of many small replicated DNA fragments called Okazaki fragments
Hydrogen bonding is still occurring when the DNA polymerase moves from a 5' to 3' direction behind the helicase that is breaking the H-bonds - this is the leading strand because its able to continually replicate because the helix is in front of it
Termination
It takes 8 hours for DNA to be replicated
At the end of DNA replication the RNA primer are replaced by DNA by 5’-3’ exonuclease and polymerase activity of ε DNA polymerase.
Exonuclease activity of DNA polymerase removes the RNA primer and polymerase activity adds dNTPs at 3’-OH end preceding the primer.
In eukaryotic organism with linear DNA, there is a problem.
When RNA primer at 5’ end of daughter strand is removed, there is not a preceding 3’-OH such that the DNA polymerase can use it to replace by DNA. So, at 5’ end of each daughter strand there is a gap (missing DNA). This missing DNA cause loss of information contain in that region. This gap must be filled before next round of replication.
Enzymes that finish replication
Gyrase (a topoisomerase)
telomerase
Gyrase (a topoisomerase)
relaxes supercoils produced when the molecule is twisted during replication. Also facilitates unwinding at beginning of replication.
Telomerase
RNA-dependent DNA polymerase
uses a short RNA template to add short DNA repeats to the short ends of linear chromosomes when the last primer is removed using RNA template
Telomeres get shorter by 10 to 15 bases each replication so older people are at a higher risk of age-related diseases due to shorter telomeres
regulatory proteins
don’t code but regulate expression and stability
RNA
Ribose has a hydroxyl group on 2nd carbon instead of hydrogen like DNA
Hydroxyl group makes more prone to degradation because phosphodiester bond is less stable than in DNA
DNA is very stable whilst RNA is not partially due to differences in sugars
RNA composition
A, G, C, U
Uracil - Demethylated thymine = less stable
Less resistant to photochemical mutations
properties of RNA
single-stranded - 5' end up into a loop down to 3' end. The stem will have comp pairing between bases that aid stability and allow RNA to remain in nucleus for enough time. Allows RNA to be less vulnerable to degradation
Read from 5' to 3'
RNA sequence comes from its DNA
Carries genetic info
Always shorter than DNA in eukaryotes
Can form hairpin structures so that it is stable for enough time for the protein to be expressed
Gene codes, RNA expresses
tRNA
Transfer RNA: Brings amino acids to ribosomes during translation.
4 loops - conserved 3 loops and free loop
are encoded by tRNA genes.
All tRNA molecules are similar in size and shape.
have CCA at the 3' end to which the amino acid attaches
At the other end is the ANTICODON, which, during translation, "reads" the matching codon on the mRNA.
mRNA
Messenger RNA: Encodes amino acid sequence of a polypeptide.
Single RNA strand
4 domains- size depends on transcript length of gene
Transcription summary
DNA unravels to be accessible to transcriptional machinery
Helicase and topoisomerase keeps DNA in open confrontation for transcription
RNA polymerase read code and make comp template pre-mRNA using other stand as template
Pre-mRNA - transcript of whole coding region
Pre-mRNA is processed via RNA processing = shorter mRNA
All of this is in nucleus
mRNA exported out of nuclear pore into cytoplasm. Then trafficked to RER where ribosomes are and is translated
Whole processes also in mitochondria for mitochondrial DNA
4 types of RNA
If infidelity in any RNA form can lead to mutation with detrimental effect due to them being fundamental genes within eukaryotic cell
mRNA
tRNA
rRNA
snRNA
rRNA
Ribosomal RNA: With ribosomal proteins, makes up the ribosomes, the organelles that translate the mRNA.
snRNA
Small nuclear RNA: With proteins, forms complexes that are used in RNA processing in eukaryotes. (NOT found in prokaryotes.)
A protein-coding gene
consists of a PROMOTER followed by the CODING SEQUENCE for the protein and then a TERMINATOR.
promoter
a base-pair sequence that specifies where transcription begins
at 5' end
Variable in sequence with some highly-conserved elements
Has transcriptional start site - allows transcription factors and RNA polymerase to bind
coding sequence
a base- pair sequence that includes coding information for the polypeptide chain specified by the gene.
Mutations due to errors in DNA replication or DNA exposure to mutagens can have detrimental or advantageous effects if within crucial regions like promoter or coding sequence
terminator
a sequence that specifies the end of the mRNA transcript
Transcriptional end site
TRANSCRIPTION- BIOSYNTHESIS OF mRNA
RNA Polymerase recognises the PROMOTER and begins TRANSCRIPTION Pre-mRNA is synthesised in a 5’ to 3’ direction
As the Polymerase moves along the DNA template the RNA is released and the DNA helix reforms
Once the Polymerase hits the terminator sequence it STOPS and the Full Pre-mRNA transcript is released.
In order for RNA polymerase to work in a 5' to 3' direction, it must use the parent 3' to 5' strand as its template
coding strand
not used as a template, but is identical in sequence to the mRNA except that all the U's are T's
template strand
what is used as a template in the synthesis of mRNA
mRNA in Prokaryotes
The sequence of a prokaryotic protein-coding gene is colinear with the translated mRNA.
The transcript of the gene is the molecule that is translated into the polypeptide.
No RNA processing happens - it has the same structure of the gene it has transcripted
mRNA in Eukaryotes
The sequence of a eukaryotic protein-coding gene is typically NOT COLINEAR with the translated mRNA.
The transcript of the gene is a molecule that must be PROCESSED to REMOVE these extra sequences called ‘INTRONS’ BEFORE it is translated into the polypeptide.
RNA processing - adds additional stability to RNA. It is processed
Both ends of a eukaryotic pre-mRNA molecule are modified by enzymes during transcription, and these modifications remain in the mRNA that is produced
At the 5' end, a cap is added consisting of a modified GTP (guanosine triphosphate). This occurs at the beginning of transcription. The 5' cap is used as a recognition signal for ribosomes to bind to the mRNA.
At the 3' end, a poly(A) tail of 150 or more adenine nucleotides is added. The tail plays a role in the stability of the mRNA - Allows it to be within the nucleus and cytoplasm for longer
pre-mRNA
Most eukaryotic protein-coding genes contain segments called introns, which break up the amino acid coding sequence into segments called exons. The transcript of these genes is the pre-mRNA (precursor-mRNA).
The pre-mRNA is processed in the nucleus to remove the introns and splice the exons together into a translatable mRNA.
Pre-mRNA Processing (Splicing)
The intron loops out as snRNPs (small nuclear ribonucleoprotein particles, complexes of snRNAs and proteins) bind to form the SPLICESOME
The intron is excised, and the exons are then spliced together.
The resulting mature mRNA may then exit the nucleus and be translated in the cytoplasm.
Lysine
Highly positively charged A.A and important for condensing DNA information into a structure within the nucleus that allows it to be compacted and also allows it to be opened up when it is require to be replicated or transcribed
Start codon
AUG = Methionine
stop codon
UAA, UAG. UGA
degenerate
In most cases more than one codon per amino acid (max. 6)
Partial DEGENERACY - first two nucleotides are identical but the third (i.e., 3′ base) nucleotide of the degenerate codon differs
complete DEGENERACY - any of the 4 bases can take third position and still code for the same amino acid
How do we READ the RNA and make a peptide
1) Once a gene has been sequenced we need to identify the OPEN READING frame
2) Every region of DNA has SIX possible reading frames (three in each direction)
BUT remember DNA is double stranded
ATG when transcribed = Aug = MET = start codon
Have to write N-Met to show start
C has to be written to show stopping
read from 5' to 3' at all times
open reading frame (ORF)
run of codons that starts with ATG and ends with a termination codon, TGA, TAA or TAG.
promoter
regulatory region of DNA located upstream (towards the 5' region) of of a gene, providing a control point for regulated gene transcription
enhancer
short (50–1500 bp) region of DNA that can be bound by proteins –ACTIVATORS to > the transcription of a particular gene
TATA box
a sequence of DNA, consisting of nucleobases TATAAA, located in the promoter region about 25-30 base pairs before the site of transcription
Most promoter regions of genes do not contain a TATA box. In TATA-less genes, transcription factors recognize other promoter sequences and RNA polymerase binds to these instead.
motif
allows identification of important region for transcription. In genes that don’t have tata box
ribosomes
the organelles on which the mRNA is translated, consist of two subunits, each of which contains rRNA and ribosomal proteins.
large Subunit [60S] - 28S rRNA, 5.8S rRNA, 5S rRNA + ~50 ribosomal proteins
Small Subunit [40S]- 18S rRNA + ~ 30 ribosomal proteins
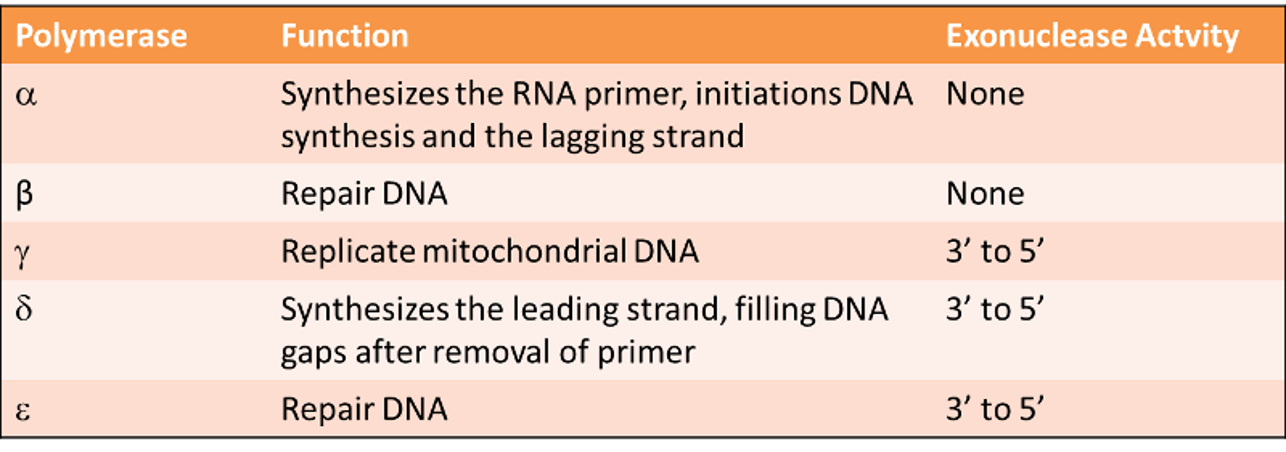
Adding an Amino Acid to tRNA
AMINOACYLATION or ‘CHARGING’- An enzyme called AMINOACYL-t RNA SYNTHETASE adds the correct amino acid to its tRNA.
Since there are 20 amino acids, there are 20 aminoacyl- tRNA synthetases.
All tRNAs with the same amino acid are charged by the same enzyme, even though the tRNA sequences, including anticodons, differ.
tRNAs are identified by their synthetases by contacts that recognize a small number of bases, typically from 1-5.
Often one of the last three base pairs in the acceptor stem is recognized. An extreme case is represented by alanine tRNA, which is identified by a single uniqu base pair in the acceptor stem.
Usually (but not always), at least one base of the anticodon is recognized. Sometimes all the positions of the anticodon are important.
Elongation of the Polypeptide Chain
Elongation of the polypeptide chain begins by the appropriate aminoacyl-tRNA binding to the codon in the A site of the ribosome.
Termination of Translation
At a stop codon, a release factor reads the triplet, and polypeptide synthesis ends; the polypeptide is released from the tRNA, the tRNA is released from the ribosome, and the two ribosomal subunits separate from the mRNA.
Polysomes
Several ribosomes can translate a mRNA at the same time, forming a polysome.
More than one ribosome can translate a mRNA at one time, making it possible to produce many polypeptides simultaneously from a single mRNA
Mismatch repair
When an incorrect nucleotide is added to the growing strand, replication is STALLED by the fact that the nucleotide's exposed 3′-OH group is in the "wrong" position.
proof-reading
Gamma and delta polymerase proofread the whole of the genome
Gamma is the only polymerase in mitochondria - has to do both functions and give a daughter cell that has high fidelity to the parent
fixes about 99% of these types of errors, but that's still not good enough for normal cell functioning.
strand-slippage
Can get permanent changes in DNA due to wobble-induced/replication error leading to strand-slippage
where
As part of gene, there is the 5' UTR which will be transcribed but not translated as the first ATG needs to be found
Introns are excised during RNA maturations
Spaces of bases between 1 gene and the next. Intergenic means its not coding for a gene
If you get mutations within red arrows, pottentail problems may occur
If theyre upsteam (green ones), in the intergeneic regions, there may not be a problem in that protein
However if its within the promoter region or within very specific sites between the zone and the intron borders, then it could have a very big effect on the expression of that particular gene
Pol II promoter elementa
This is important because if there is a mutation within the promoter, it will be dependent on where in the promoter it has happened
If its within the intergenic regions between the tata and CAAT box, the chances of it having a big effect is minimal
However, if its in one of those elements, it has a greater chance of having an impact on whether that gene will be transcribed
If the mutation is at the transcriptional start size, there is a 100% chance that it will affect the gene expression of that gene
induced physical damage to DNA
Environmental agent(s)
Exposure to chemicals UV rays
spontaneous physical damage to DNA
without any exposure to any environmental agent
Spontaneous biochemical reactions taking place within the cell
Wobble-induced/replication errors
point mutations
single base pair substitutions
transition
transversion
transition substitions
Purine REPLACED BY A Purine
Adenine = Guanine
Pyrimidine REPLACED BY A Pyrimidine
Thymine = Cytosine
Transversion substitions
purine being replaced by a pyrimidine pyrimidine being replaced by a purine
A/T = G/C
silent mutations
no change in the protein sequence
missense mutations
change the amino acid sequence
protein is okay
nonsense mutation
Introduces a stop codon to early
frameshift missense mutation
A base is excised from the parent strand = produced RNA is one base shorter than parent
Everything has shifted so codons have changed = frame shift
Original stop codon has been removed so it has to carry on until another is found = different protein to the one that was suppose to be made
packaging
DNA is packaged into chromatin
The DNA fundamental unit of chromatin is the nucleosome
Nucleosome, the fundamental unit of chromatin, consists of histones
histones
The MOST common NUCLEAR proteins.
Account for almost half of the proteins isolated from nuclei.
Histones broken down into DNA, which is what is being packaged, proteins, which are going to form part of the nucleosome and a small amount of non-coding RNA which helps keep that transcriptional regulation of genes in play
1:1 ratio of amount of histones to DNA mass that’s present
Non-histone components needed to allow high-level packing of DNA
histone family of proteins
Histones are small and are highly positively charged. Also highly conserved
Very highly conserved histones have a very significant role within cell
H3 & H4 are two of the most highly conserved proteins that a eukaryotic cell has
Followed closely by H2A and H2B, which are slightly more diverse
but still highly conserved compared to other crucial enzymes in nucleus
H1 shows more divergence but yet still conserved
DNA double helix
NA does not exist as helix in nucleus
Only when acts as a linker between adjacent nuclear sites
DNA is packaged at all times
From 10nm fibre to 30nm fibre up to 300 through to 700
Highly compacted chromatin when we hit metaphase and then the splitting of chromosomes so that we can transfer the genetic information from one cell generation to the next
So by being highly packaged, the genome is protected during the mitotic phase
basic unit of chromatin is the nucleosome
FOUR CORE Histones: H2A, H2B, H3, H4 - Make up nucleosome, 2 molecules of each which make up central optometric core
ONE LINKER Histone: H1 - 1 molecule that is associated with octameric core
146bp of DNA LEFT HANDED SUPERHELIX. DNA path = 1.8 superhelical turns Wrap around nucleosome
Level 1 of DNA Packaging: The Nucleosome
146 base pairs wrap around octamer, histone's central core
And then DNA needs to link from one nucleosome to the next
= around 200 base pairs
Mass of DNA : mass of protein
Heterodimerization of histones H3 and H4
Structure causes histone fold. They fold together and clamp onto each other to make 'handshake' motif, which is very specific to the nucleosome
2 molecules of H3 and H4 which associate together = 2 pairs of 'handshakes' - join together in centre of optometric core to make horseshoe shape in the centre of the nucleus
Histones H2A and H2B form a Dimer
A pair of H2A and H2B binding together in 'handshake' but this time the nucleosome of H3 and H4 are in the centre so H2A and H2B bind above and below
This gives an optometric core around which DNA is wrapped in 1.8 super helical turns and packaging 146 base pairs
If a histone should ‘be altered’ within the octamer then a change in the path of the DNA around the octamer occurs and this may result in a change in the ‘packaging’ of the DNA.
histone variants examples
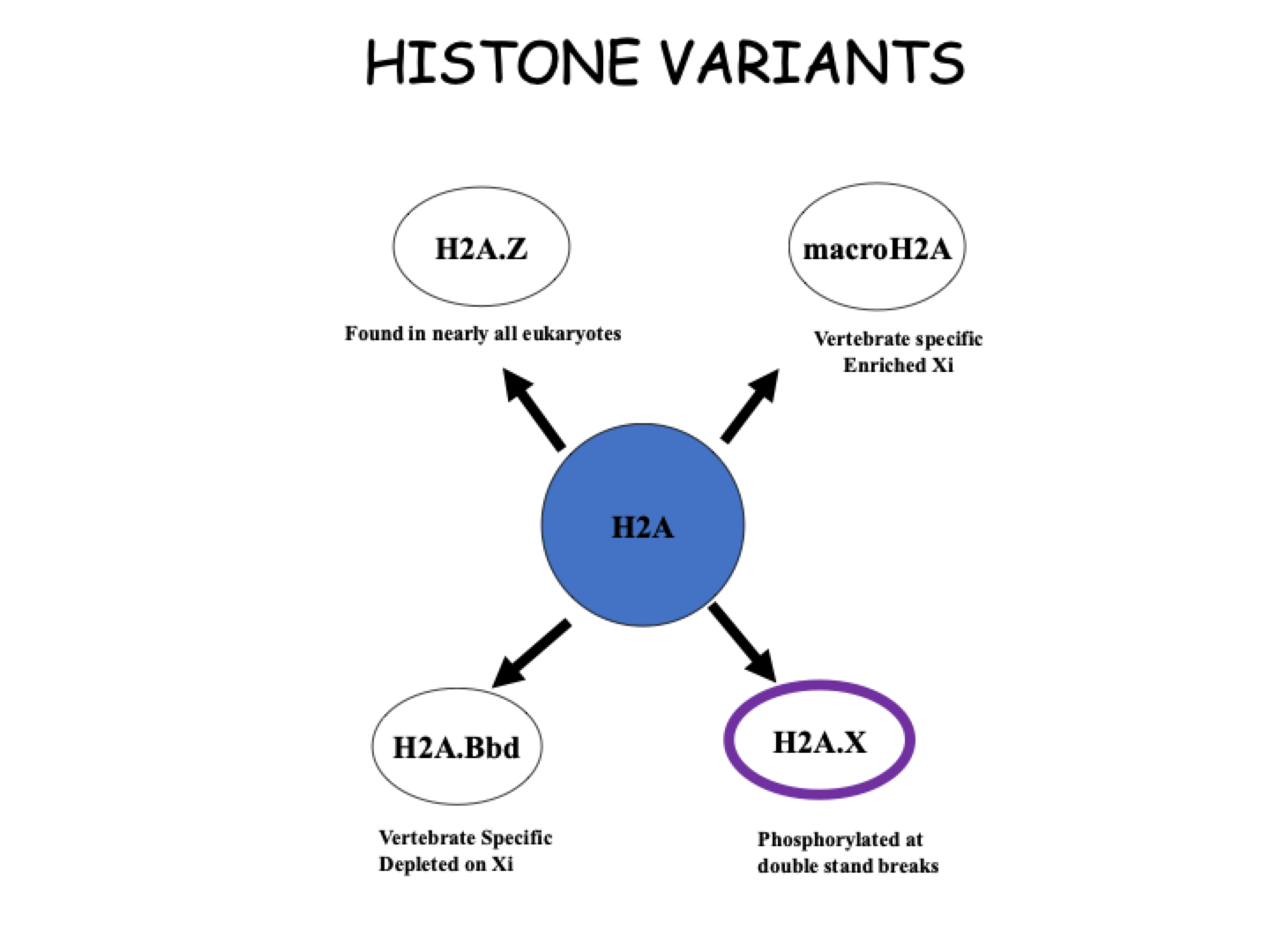
In females, one of the X chromosomes is inactivated randomly but it must remain inactive and for that to happen, it is made different frim the active X chromosome by pushing away the normal H2A and filling throigh those nucleosomes with macroH2A
When DNA is damaged, there is a break because the DNA is packaged in new Which is signified by the expulsion of the normal H2A and replaced bt H2A.X
H2A v H2A.Z
H2A.Z has only a 60% IDENTITY to H2A
Alters the interaction stability between H2A and H2B
Alters the interaction of the H2A:H2B dimer with H3:H4 tetramer - Interaction is looser which makes the octamer slightly bigger than normal so the DNA wrapped around becomes looser
ALTERs the CANONICAL NUCLEOSOME
H2A.Z containing nucleosomes often associated with transcriptionally active chromatin.
histone variants
Hardly any different in A.A sequence between variants but the subtle differences are sufficient enough to change the normal structure of the tetra in the centre
H3.3 - Important variant when changing the transcriptional process outside of replication
cenpA - Enriched in centromere and telomeres
Variants of histones H3 and H2A differentiate chromatin at CENTROMERES, ACTIVE GENES and HETEROCHROMATIN
Replacement of H3 with H3.3 MARKS actively transcribed loci by replication independent nucleosome assembly
Epigenetically silenced chromatin is enriched or depleted in an abundance of diverse H2A variants
level one packaging
the nucleosome
Four CHs + H1 packages 146bp + linker DNA = ~200bp DNA
DNA makes two turns around the core Forming a LEFT-handed super helix.
DNA path = 1.8 superhelical turns.
DNA does NOT follow a smooth Path around the nulceosome.
Accessibility of Nucleases restricted
level two - 10nm fibre
Lowest level of packing in nucleus
This is when DNA is packaged and each nucleosome is holding onto the next
Double helix is present in linker DNA but is connected to another nucleosome
PACKING RATIO: 6-7
level three - 30nm solenoid
Histone H1 is ESSENTIAL for these higher order forms of packaging
10nm is coiled to form 30nm
Requires 6 nucleosome to coil
PACKING RATIO: ~40
level four - 300nm solenoid
Each loop contains 60-100 kb of DNA tethered by nonhistone scaffold proteins
Bind each loop to scaffold
Loops are well contained and tethered at each end of the loops onto the scaffold so it can be accessed later on
The 30nm solenoid further coils probably employing a PROTEIN SCAFFOLD to condense the DNA still further.
PACKING RATIO: 680
level five - 700nm fibre
300nm coiled again - each loop is a 300nm fibre
Results in a tightly compacted DNA associated each time with nucleosomes going through different packaging levels
The next step is described as the COILED COIL and requires the loops of chromatin to coil again to form a condensed 700nm fibre.
PACKING RATIO: 10^4
level five - 700nm solenoid Chromosome
PACKING RATIO: 10^4
level 6: Metaphase Chromosome
Highest form of packaging
Visible under light microscope
Heterochromatin
Constitutive or Facultative
Highly condensed and in general transcriptionally INACTIVE
Contains no gene and present in telomeres and at the centromere
In most cells 90% of the genome is Inactive BUT only 10% is in this highly condensed form.
Restricted regions of chromosome
Repetitive DNA Sequences
Replicated LATE in S phase
constitutive heterochromatin
All cells of a given species will package the same regions of DNA into constitutive heterochromatin
Genes contained within the constitutive heterochromatin will be poorly expressed due to high packaging
remains CONDENSED most of time in all cells (e.g. Y chromosomes)
euchromatin
contains genes
Lightly stained regions of chromosomes
More ‘open’ chromatin configuration during interphase.
Replicated early in S phase
Contains both transcriptionally active and inactive genes
Differential histone modifications
ACTIVE v INACTIVE
Contains both transcriptionally active and inactive genes
faccultative heterochromatin
DNA packaged in facultative heterochromatin will NOT BE CONSISTENT within the cell types of a species
is regulated and is often associated with morphogenesis or differentiation.
gene is randomly inactivated between the cells. (e.g. X inactivation)
X-Chromosome Inactivation
A sequence in one cell that is packaged in facultative heterochromatin (genes poorly expressed) MAY be packaged in EUCHROMATIN in another cell (and the genes expressed).