Chap 12 gene transcription and RNA modification
1/127
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
128 Terms
gene
the segment of DNA used to make a functional product
----------------------
Types of functional product:
- RNA
- polypeptide
DNA replication
making DNA copies that are transmitted from cell and from parent to offspring
Transcription
the first step of gene expression where DNA sequences are copied into an RNA sequence
- the structure of DNA is NOT altered
(can continue to store info)
----------------------
- its a particular segment of DNA thats copied into RNA by the enzyme RNA polymerase
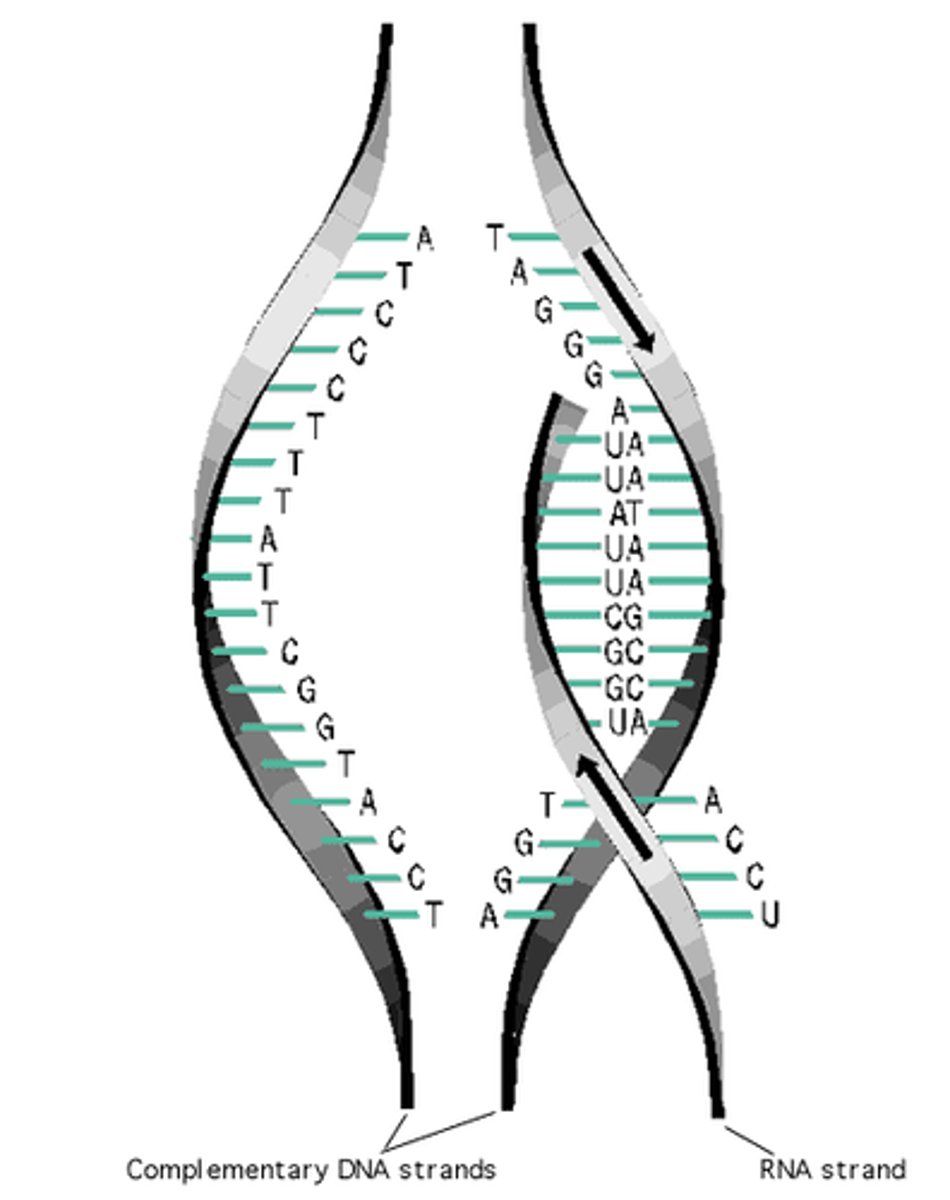
Translation
Process by which mRNA is decoded and a protein is produced
- produces an RNA copy of a gene
Directions of transcription and translation
Down stream (going right)
- 3'-5' = transcription and translation
----------------------
Up stream (going left)
5'-3' = direction from which the polymerase or ribosome has come
structural genes
AKA: protein-encoding genes
----------------------
- genes that encode the amino acid sequence of polypeptides
Chromosomal DNA
stores information in units called genes
Messenger RNA (mRNA)
the product of transcription
- these type of nucleotide sequences determines the amino acid sequence of a polypeptide during translation
Polypeptide
long chain of amino acids that makes proteins
- becomes part of a functional protein that contributes to an organism's traits
dogma of genetics
(RECAP)
DNA -> RNA -> Protein
----------------------
- DNA -> RNA = transcription (script)
- mRNA -> protein = translation (read)
----------------------
- defined as the path from gene to trait
gene expression
overall process by which the information within a gene produces its product and the product carries out its function
- describes how DNA produces a protein
Regulatory sequences (DNA)
site for the binding of regulatory proteins
- can be found in a varity of locations
----------------------
role:
- influence rate of transcription
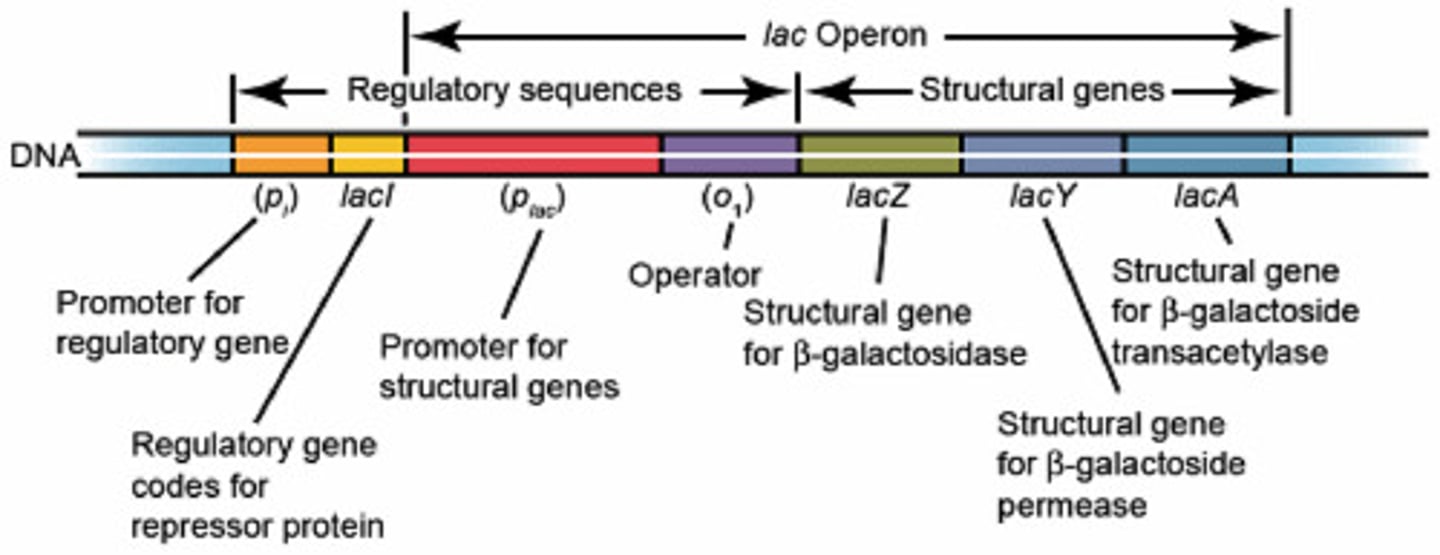
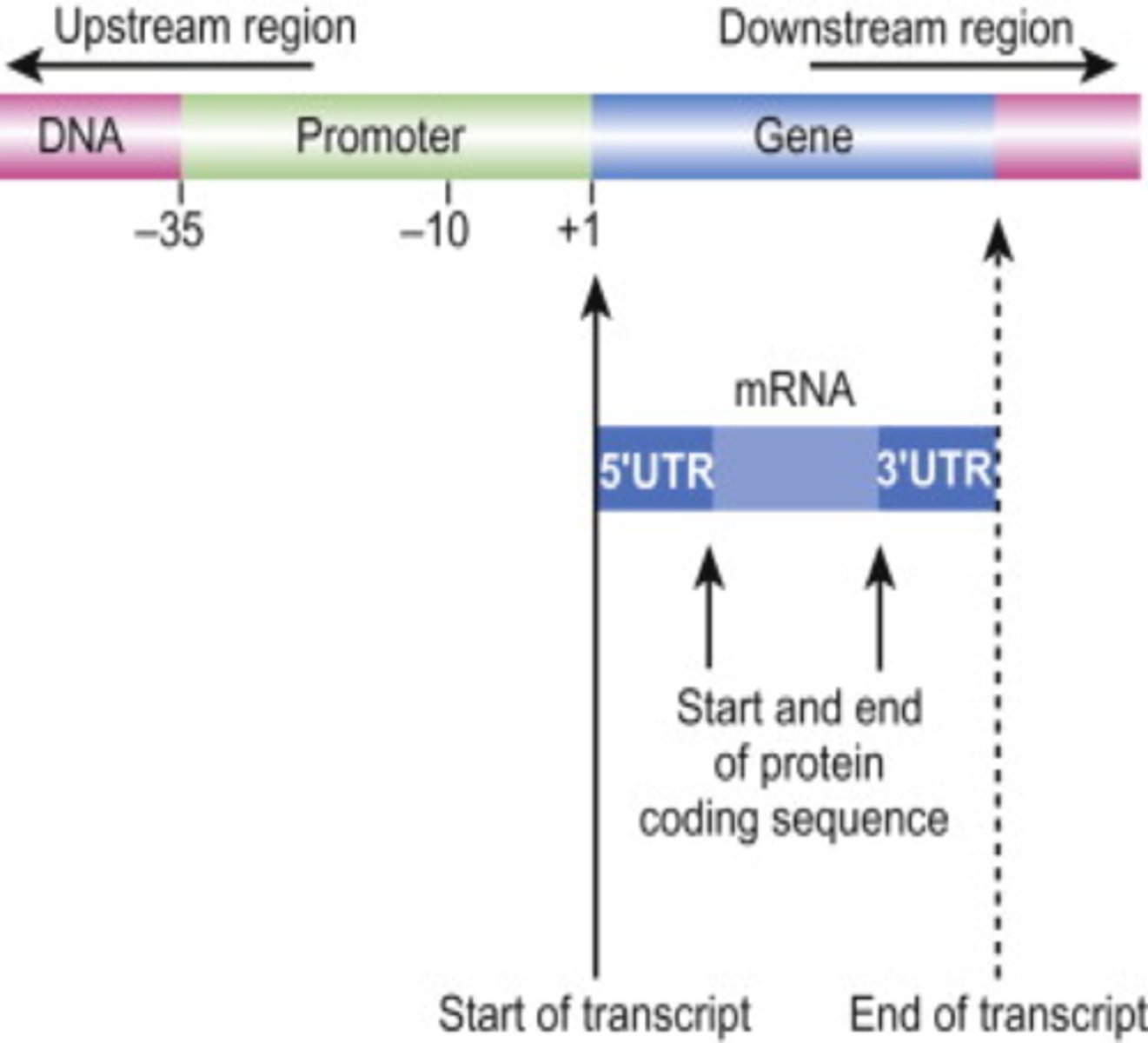
Promoter (DNA)
region of a gene where RNA polymerase can bind and begin transcription
- direct the exact location for the initiation of transcription
- located upstream of the site where transcription of a gene actually begins
- bases are #ed in relation to the transcription start site
transcription start site
The site at which the first RNA nucleotide is added
AKA +1 site (starts on either end of the promoter)
terminator (DNA)
signals the end of transcription/RNA synthesis
ribosomal binding site (mRNA)
Site for ribosome binding; translation begins near this site in the mRNA.
----------------------
In eukaryotes, the ribosome scans the mRNA for a start codon.
Codons
3-nucleotide sequences within the mRNA that specify particular amino acids.
- sequence of codons within mRNA determines the sequence of amino acids within a polypeptide.
start codon (AUG)
specifies the first amino acid in the polypeptide sequence
- Bacteria/Prokaryotes = formylmethionine
- Eukaryotes = methionine
stop codon (UAA, UGA, UAG)
specifies the end of polypeptide synthesis
----------------------
Bacteria stop codons:
- may be polycistronic
polycistronic
the encoding of two or more polypeptides via stop codon
nomenclature for types of strands
DO NOT MIX THE NAMES UP WITH ANOTHER SET
- Set 1) template strand and non-template strand
- set 2) Non-coding strand and coding strand
- set 3) Anti-sense strand and sense strand
template strand
DNA strand that is actually transcribed
- the RNA transcript will always be complementary to the this strand
- has the same base sequence as an RNA transcript (exception of T in DNA and U in RNA)
----------------------
AKA: non-coding or anti-sense strand
(DO NOT MIX SETS UP)
non template strand
the strand of DNA that is not transcribed into RNA during transcription
- is the strand that is opposite to the template strand
----------------------
Features:
- the base sequence is identical to RNA transcript
(except for the substitution of uracil in RNA for thymine in DNA)
----------------------
AKA coding or sence strand)
DO NOT MIX SETS
transcription factors
recognize the promoter and regulatory sequences to control transcription
- are usually involved in transcription purposes
----------------------
types of transcription factors:
- ribosomal binding sites
- codons
stages of transcription
1. Initiation
2. Elongation
3. Termination
----------------------
- all involve protein DNA interactions
(where proteins like RNA polymerase interact with DNA sequences)
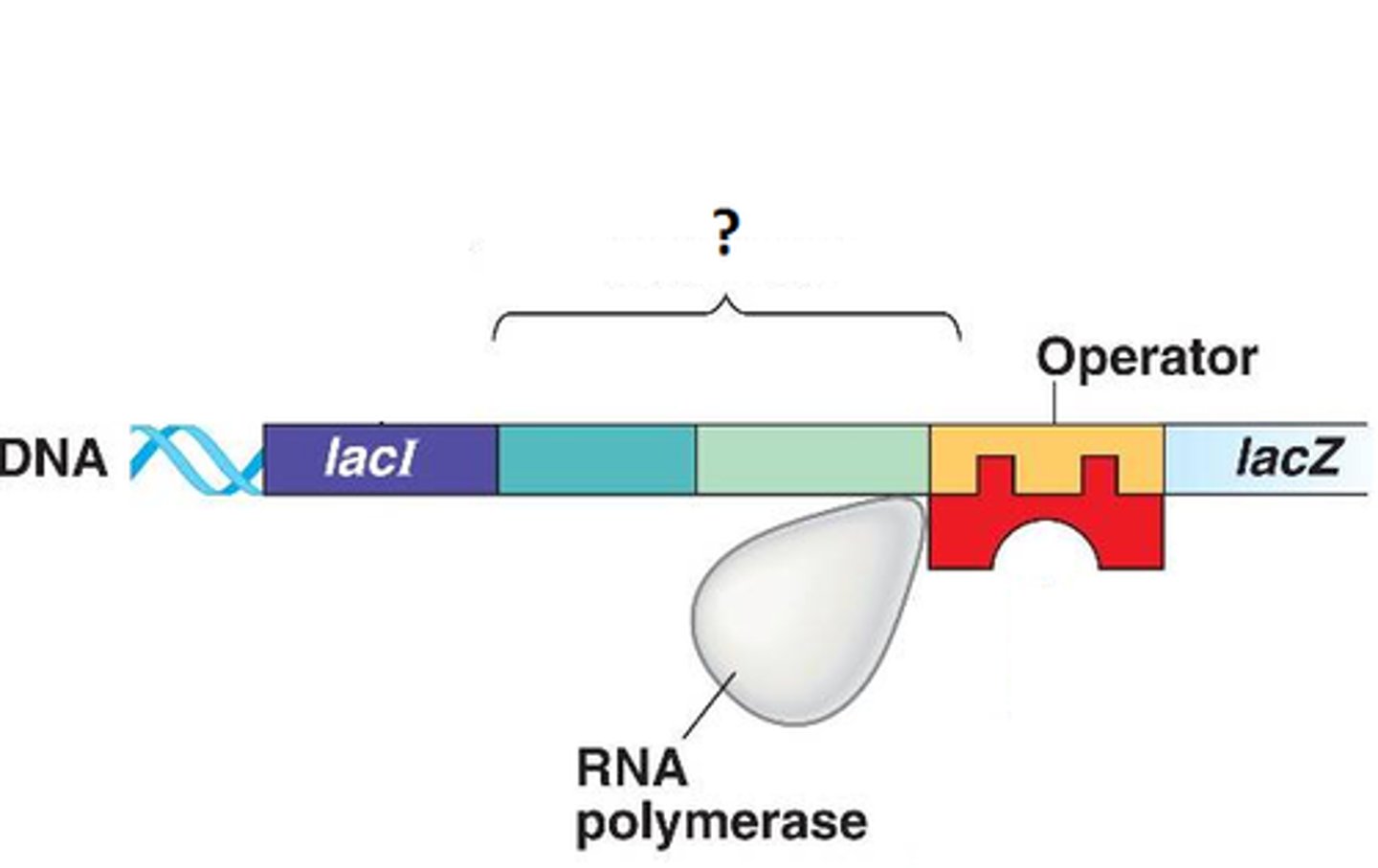
Initiation (transcription)
The promoter functions as a recognition site for transcription factors.
- the transcription factors enable RNA polymerase to bind to the promoter.
- after binding, the DNA is denatured into a bubble known as the open complex.
open complex
the region of separation of two DNA strands produced by RNA polymerase during transcription
----------------------
When does it form?
- forms when the TATAAT box in the -10 region is unwound
closed complex
the complex between transcription factors, RNA polymerase, and a promoter before the DNA has denatured to form an open complex
----------------------
when does it form?
- is usually formed when RNA polymerase binds to the promoter
Elongation/synthesis of the RNA transcript (tanscription)
RNA polymerase slides along the DNA in an open complex to synthesize RNA
- involves sigma factors
----------------------
During transcription, the sigma factor is released allowing the enzyme to slide down the DNA to synthesize an RNA strand
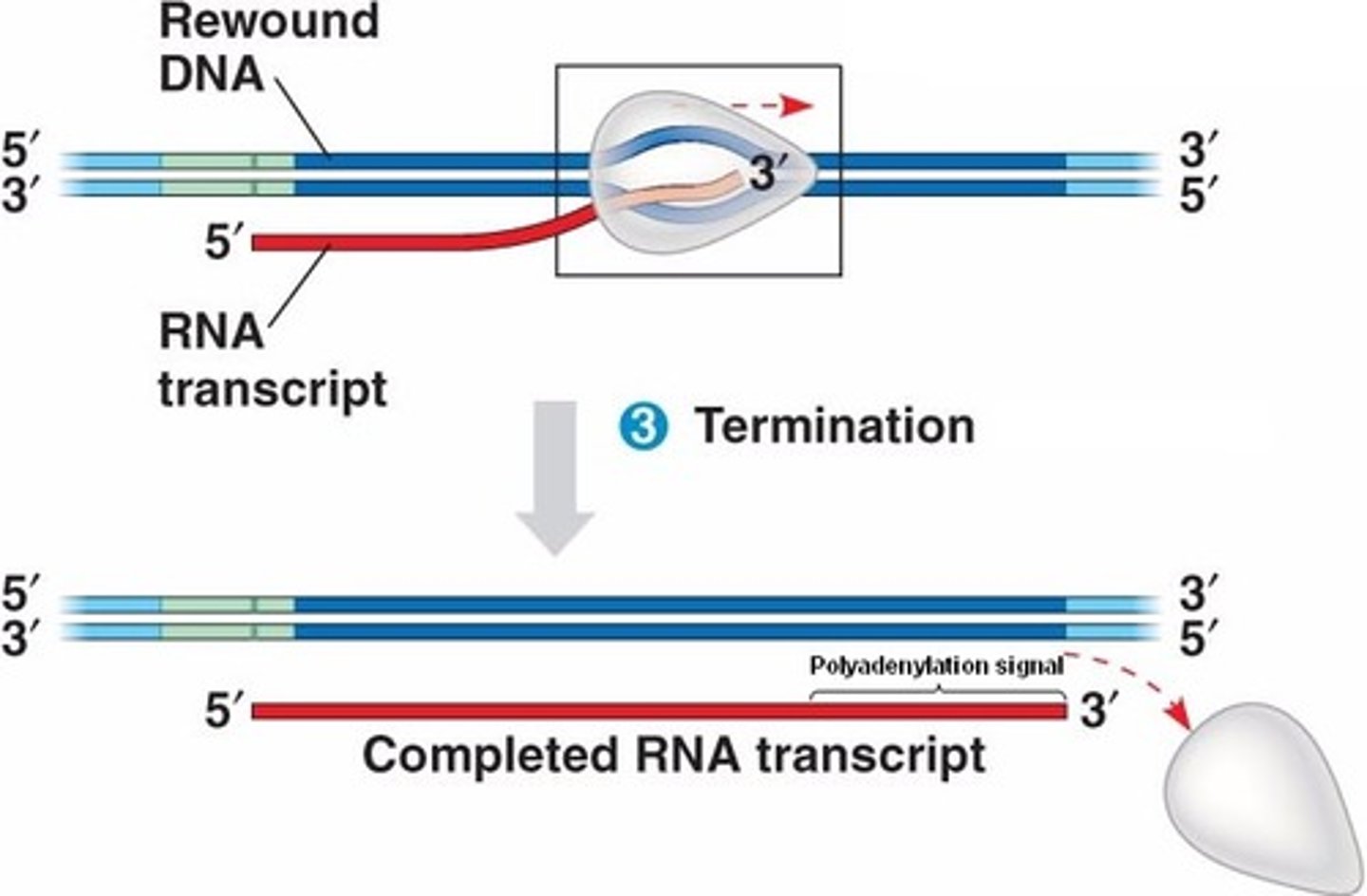
termination (transcription)
A terminator is reached that causes RNA polymerase and the RNA transcript to dissociate from the DNA
RNA polymerase
enzyme that links together the growing chain of RNA nucleotides during transcription using a DNA strand as a template
----------------------
contains 6 substrates formed into a holoenzyme
- 2 alpha subunits
- 1 beta subunit
- 1 beta prime subunit
- 1 omega subunit
- 1 sigma subunit
RNA polymerase movement
- moves along the template strand in 3'-5' direction
- RNA synthesized in a 5'-3' direction (uses nucleoside triphosphates as precursors and releases pyrophosphate)
complementary rule in RNA
A=U
G=C
Core enzyme of RNA polymerase
responsible for catalytic function
----------------------
contains 5 substrates formed into a holoenzyme
- contains every subunit from RNA polymerase except sigma subunit
sigma factor
helps the holoenzyme find a promoter for transcription
- is involved in the initiation phase for transcription
----------------------
features:
- has a helix turn helix structure (HTH)
- usually has 5'-3' polymerase activity
sigma factor special abilities
- can bind loosely to DNA
- can scan along DNA strands until it encounters a promoter region
sigma factors encountering promoter region
the sigma factor recognizes both the -35 and -10 regions
- a region within the sigma factor that contains a helix-turn-helix structure is involved in a tighter binding to the DNA
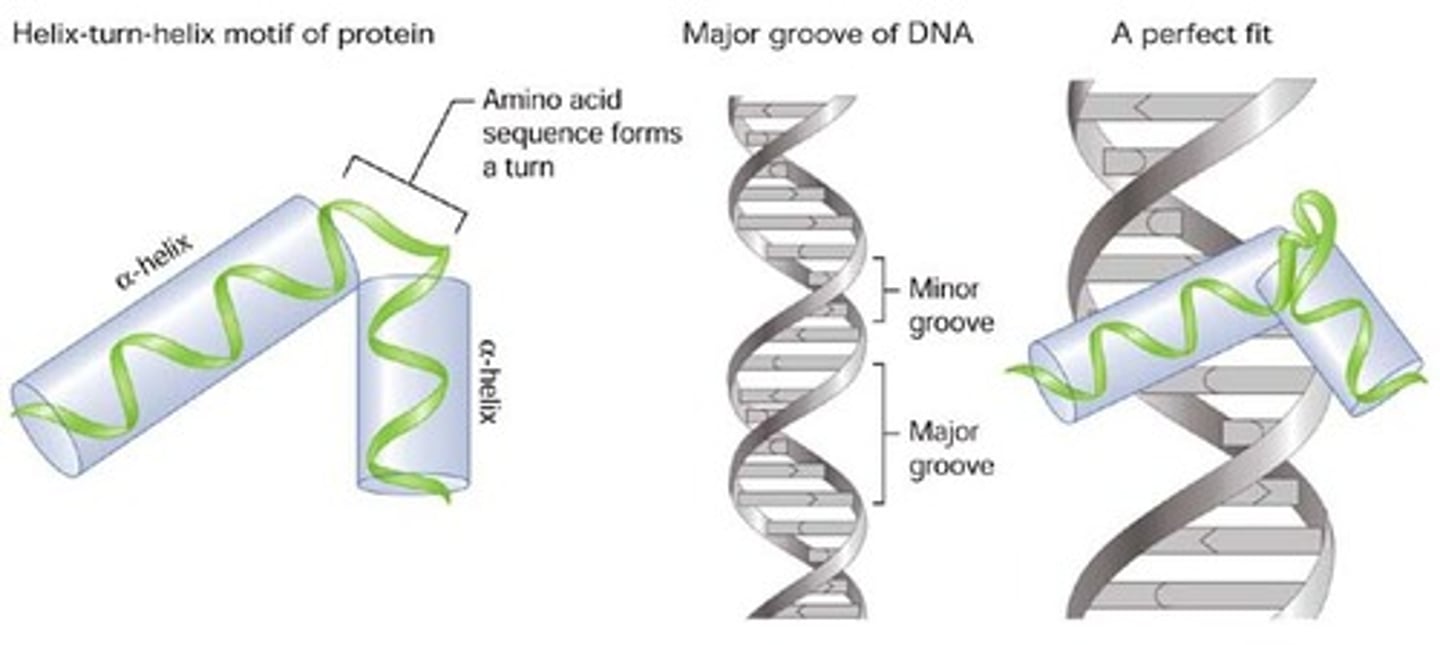
helix-turn-helix
A protein domain composed of two α helices joined by a short strand of amino acids and is found in many DNA-binding proteins.
- is responsible for recognizing the promoter
----------------------
Within sigma factor:
- fits in the major groove on the DNA strand
5' to 3' polymerase activity
adds DNA nucleotides to the 3' end of the DNA segment preceding the primer
- allows for transcription to take place
switching sigma factors in bacteria (from high to low)
Switching from one sigma factor to another in the RNA polymerase will suddenly switch off a set of genes and turn on another due to this changing the specificity of the RNA polymerase.
Deleting a promoter from a gene
if you perform a northern blot you wouldn't see any bands being formed as you would have deleted the promoter (necessary for transcription)
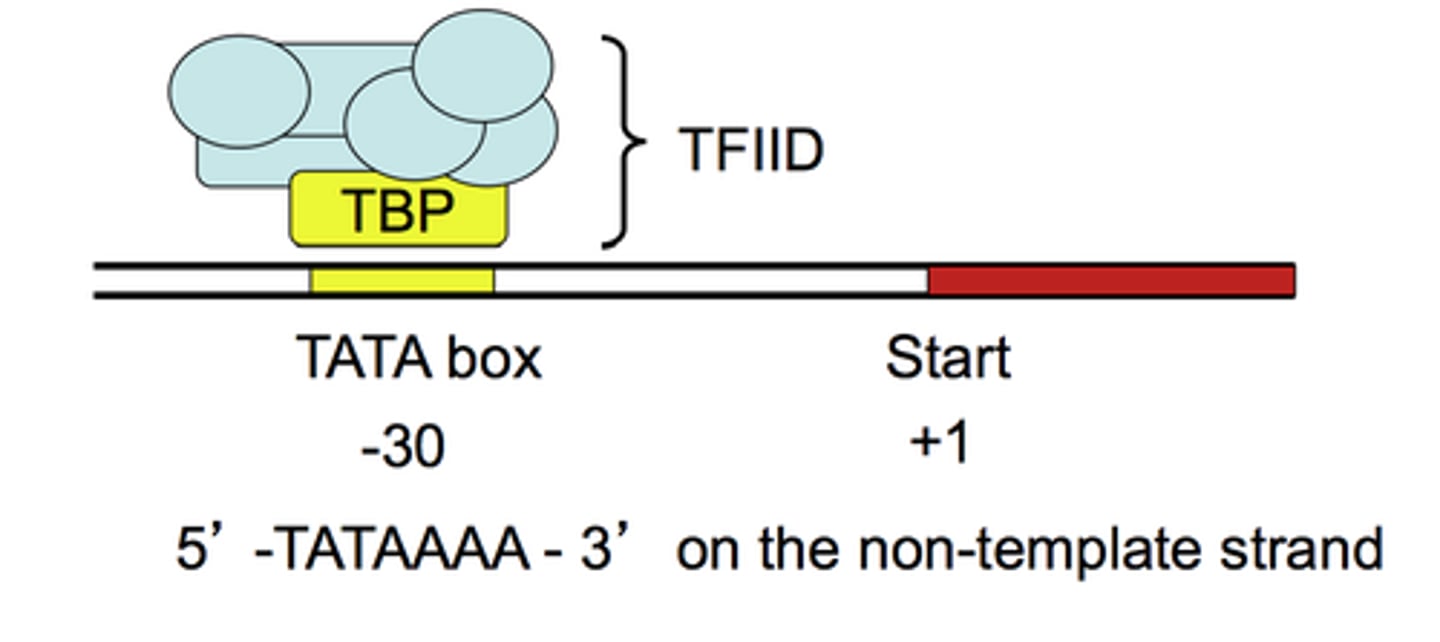
TATA box
A promoter DNA sequence crucial in forming the transcription initiation complex.
----------------------
How can alter the sequence
- you could change the TATA box by performing site-directed mutagenesis
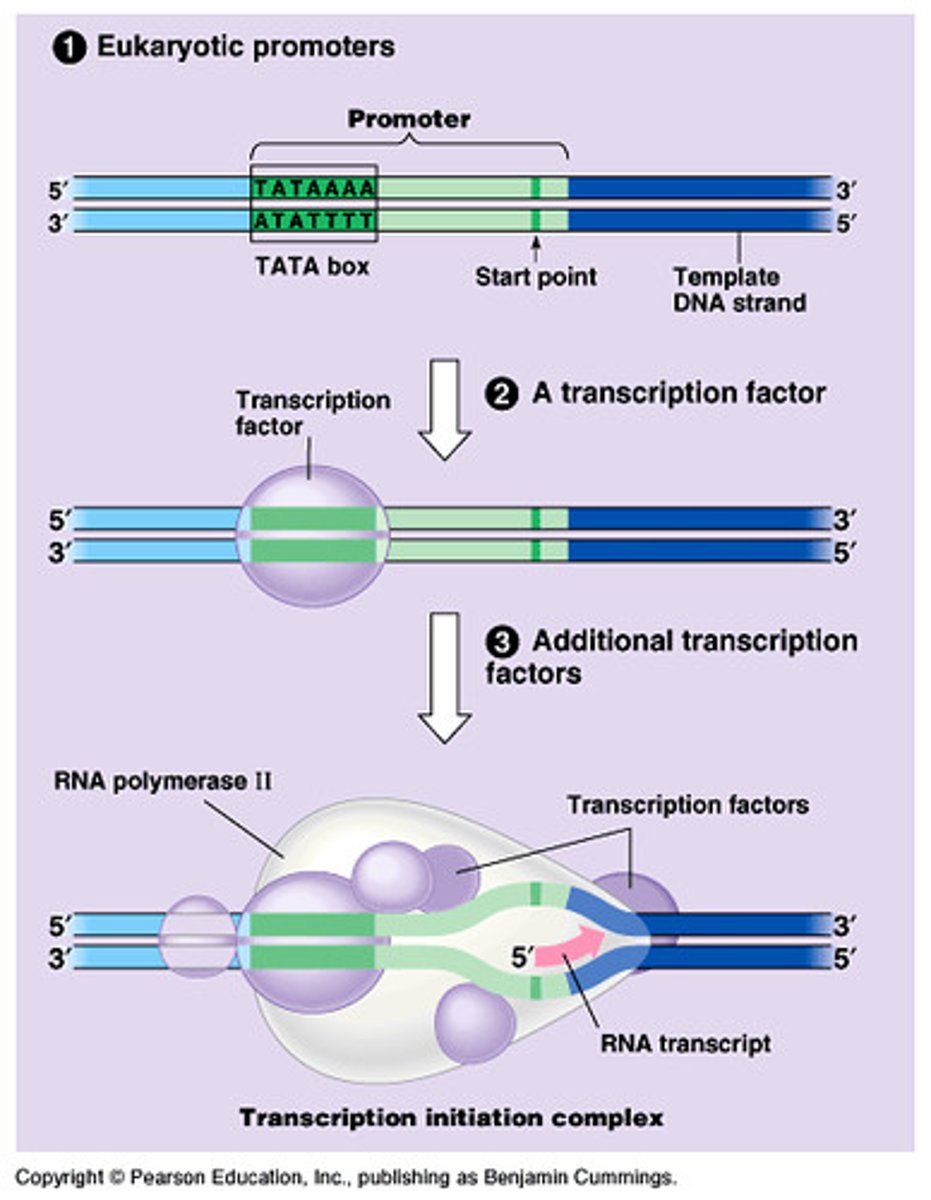
Formation of open complex
is formed by the action of RNA polymerase (17 bases long)
----------------------
What happens behind the open complex?
- DNA rewinds back into a double helix
RNA synthesis
another name for transcription
----------------------
how fast does it move?
- 43 nucleotides per second
When does termination occur?
happens when the Short RNA-DNA hybrid of the open complex is forced to separate
- this will release newly made RNA as well as RNA polymerase
Bacterial mechanisms for termination
- rho-dependent termination
- rho-independent termination
Rho-dependent termination
helicase protein AKA rho-protein terminates transcription
----------------------
rho-helicase binds to rut site and moves to the polymerase enzyme where it terminates the transcription
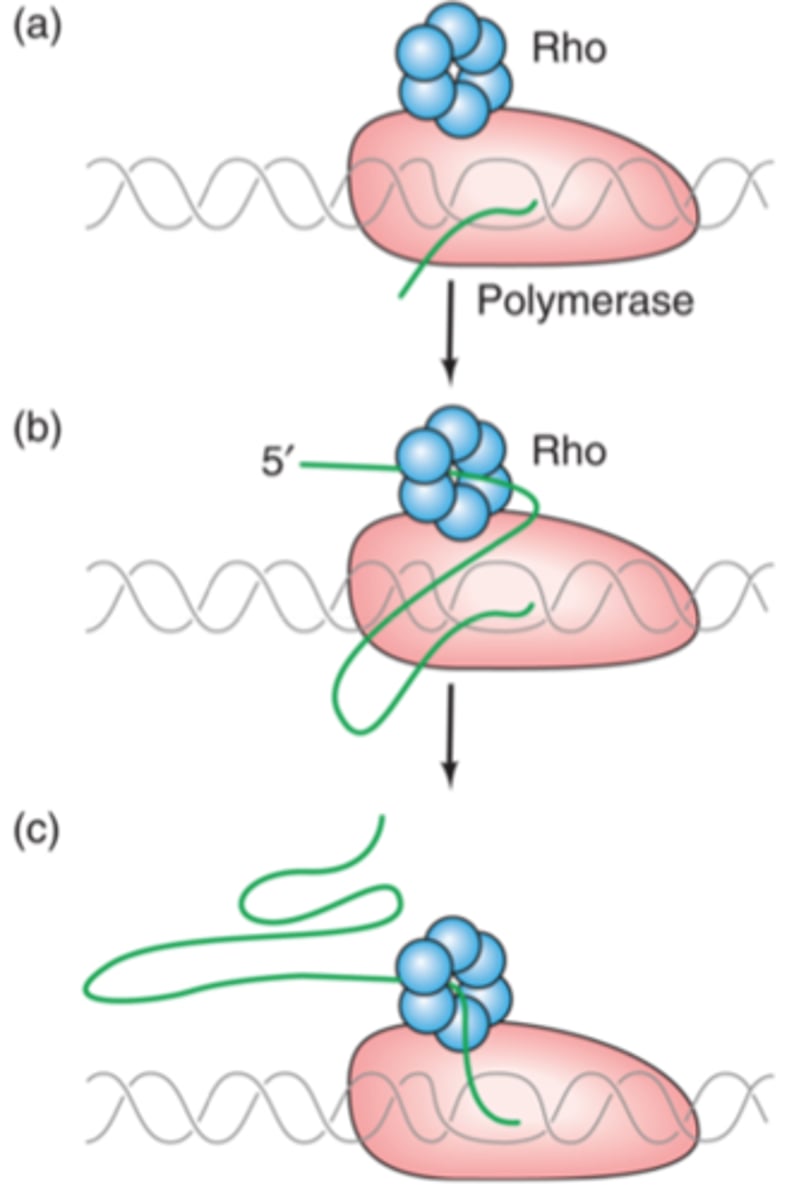
rut site
AKA: rho utilization site
- area where Rho-helicase binds towards the RNA polymerase (via ATP hydrolysis)
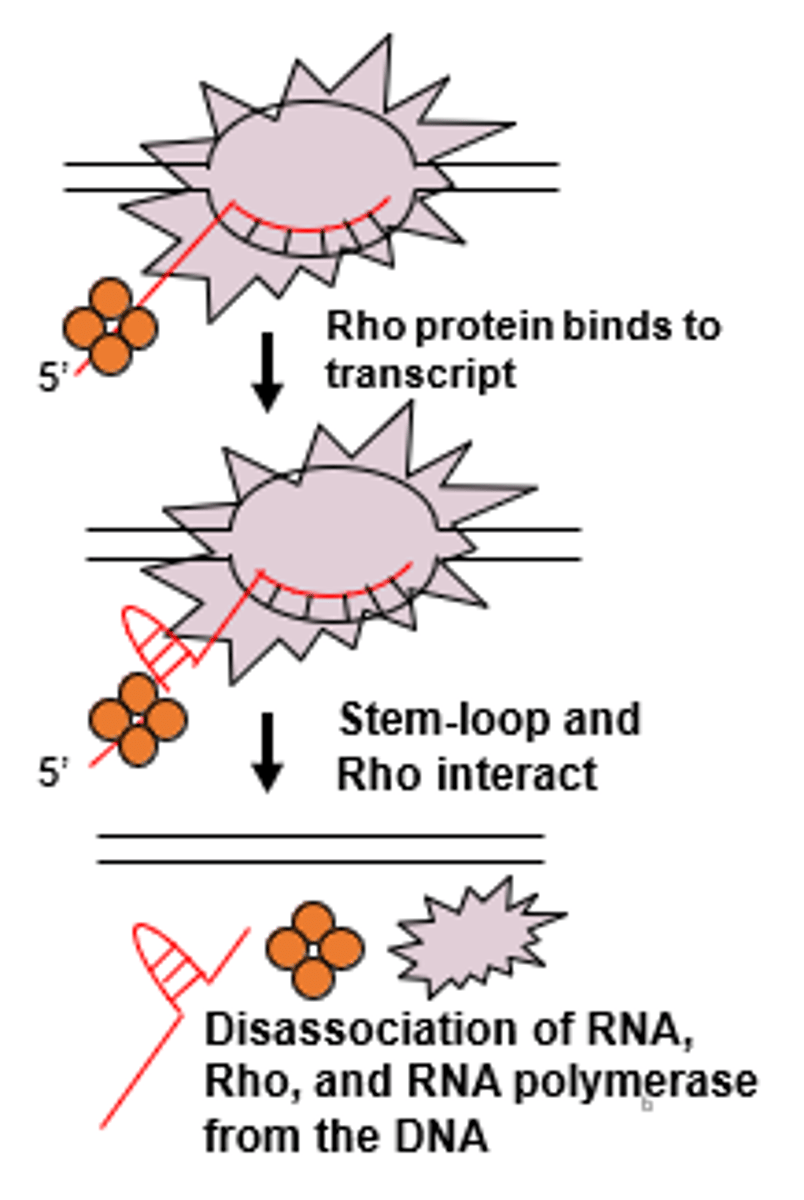
rho-dependent termination steps
1. helicase protein binds to Rut site binds
----------------------
2. helicase protein starts to move towards RNA polymerase (via ATP hydrolysis)
----------------------
3. when helicase reaches RNA molecule it encounters DNA-RNA duplex (allowing for the enzyme to unwind and the net transcript is released from the complex)
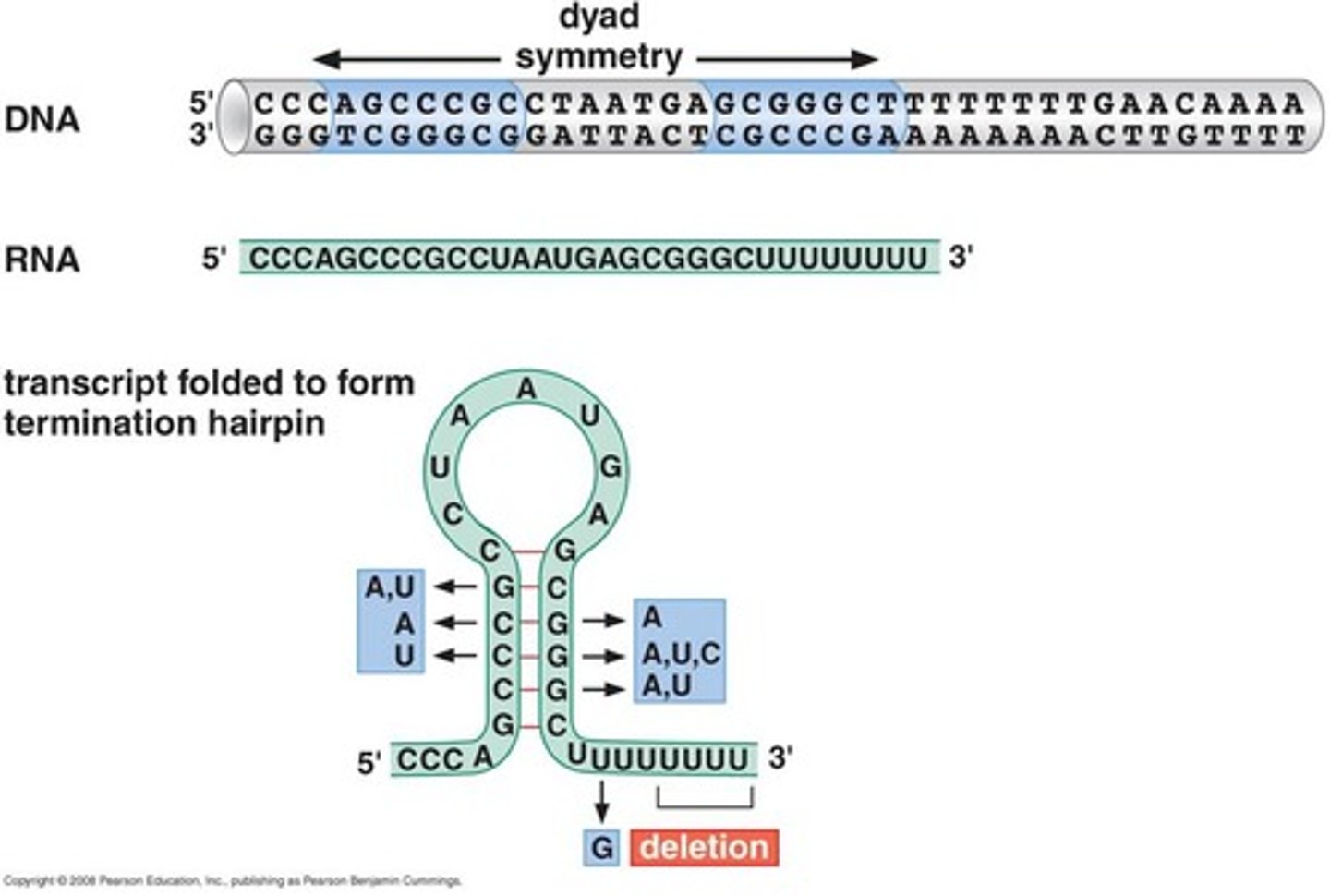
Rho-independent termination
physical modified structure of RNA transcript terminates transcription
----------------------
stem-loop structure pulls off RNA transcript from the RNA polymerase complex thereby terminating the transcription
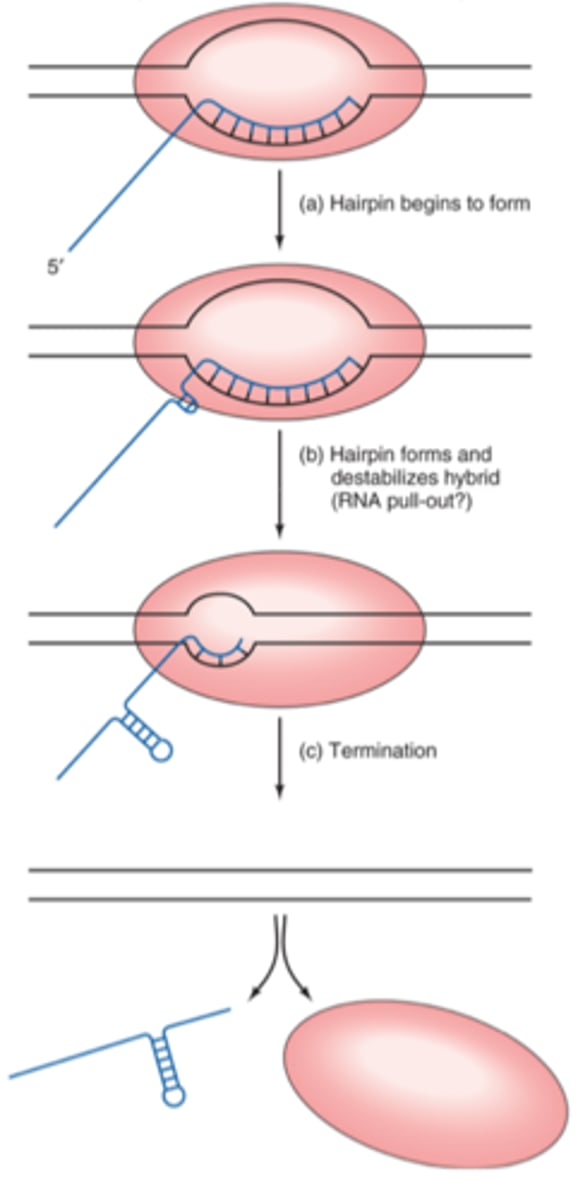
rho-independent termination steps
1. RNA transcript is synthesized on the template strand by (RNA poly.)
----------------------
2. G-C-rich regions on RNA transcript forms stem-loop structure (due to palindromic sequences on RNA transcript)
----------------------
3. stem-loop (with a uracil rich sequence) will reduce the length of the RNA-DNA hybrid allowing for physical traction to occur
----------------------
4. A-T rich regions forms a weak link within DNA strand allowing for the termination of transcription
basal transcription factors
proteins that interact with the promoter and are not restricted to particular genes or cell types
----------------------
involves 3 categories of proteins:
- RNA polymerase II
- general transcription factors (GTFs)
- mediator
RNA polymerase II
enzyme that catalyzes the linkage of nucleotides in 5'-3' direction using DNA as a template
- subunits usually wrap around it
----------------------
in eukaryotes:
- RNA poly. II proteins are composed of 12 subunits
general transcription factors
- TFIID
- TFIIB
- TFIIF
- TFIIE
- TFIIH
TFIID
Composed of TATA-binding protein (TBP) and other TBP-associated factors (TAFs). Recognizes the TATA box of eukaryotic protein-encoding gene promoters.
TFIIB
Binds to TFIID and then enables RNA polymerase II to bind to the core promoter. Also promotes TFIIF binding
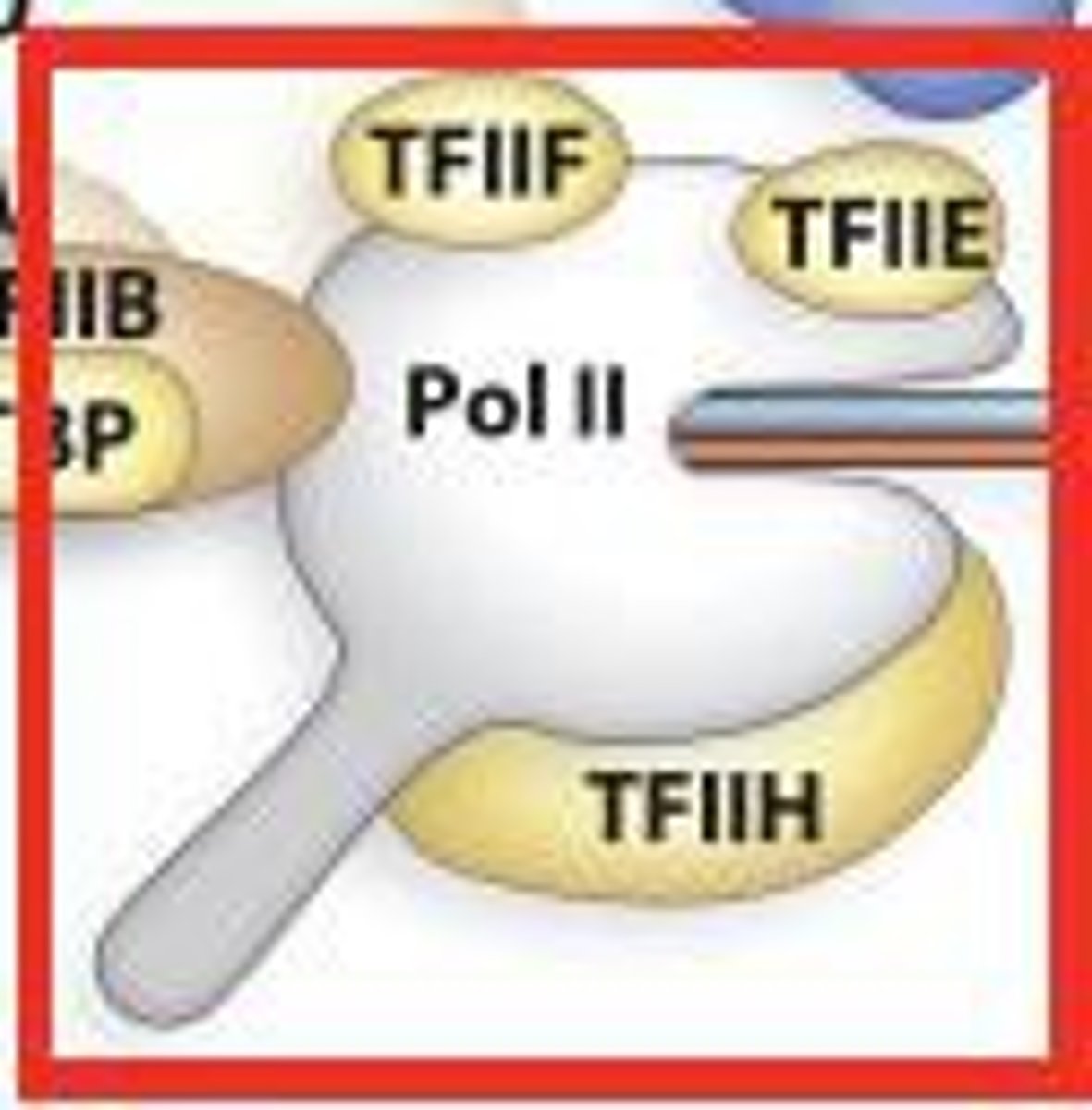
TFIIF
binds to RNA polymerase II and plays a role in its ability to bind to TFIIB and the core promoter
----------------------
plays a role in the ability of TFIIE and TFIIH to bind to RNA polymerase II
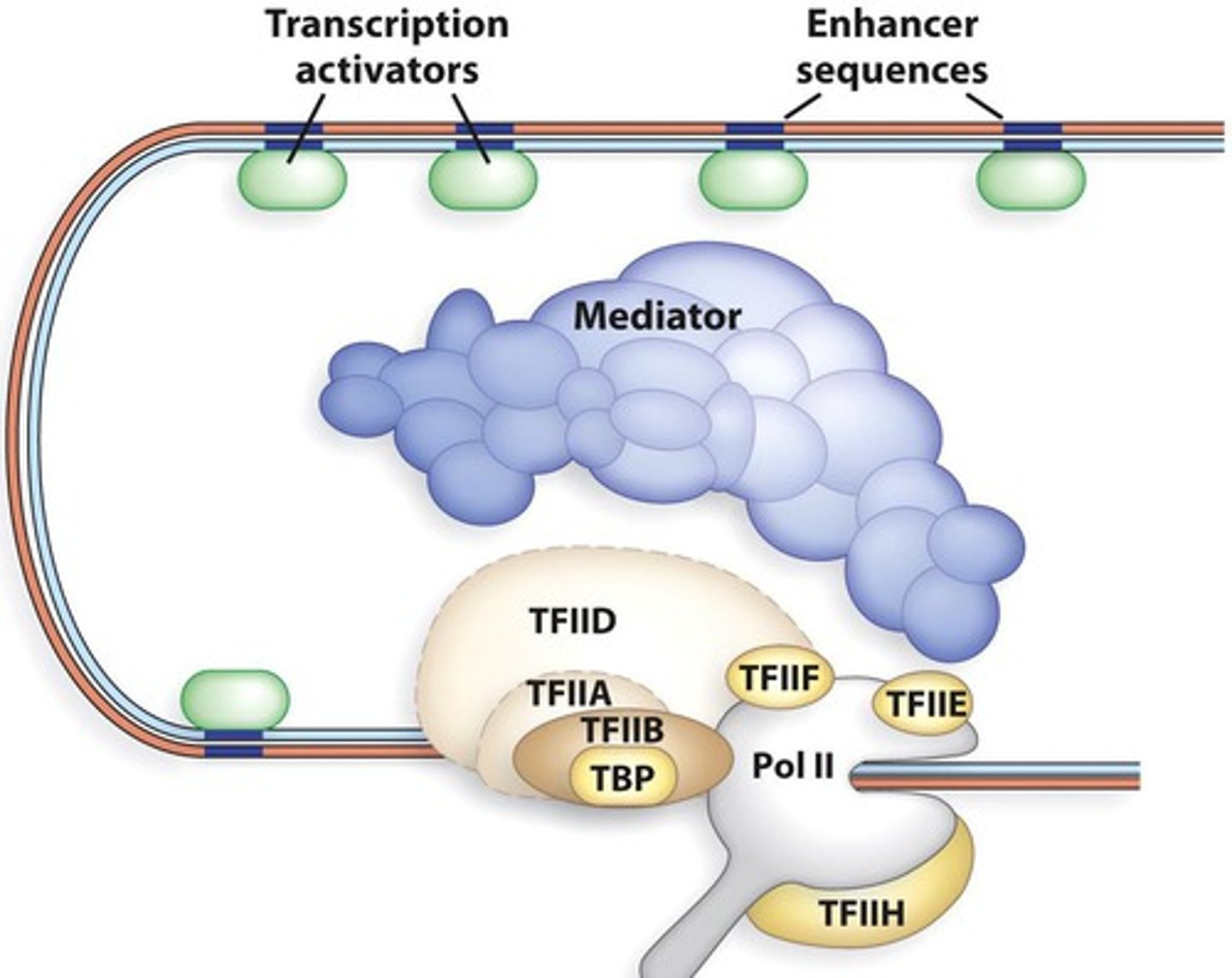
TFIIE
Plays a role in the formation or the maintenance (or both) of the open complex.
----------------------
It may exert its effects by facilitating the binding of TFIIH to RNA polymerase II and regulating the activity of TFIIH.
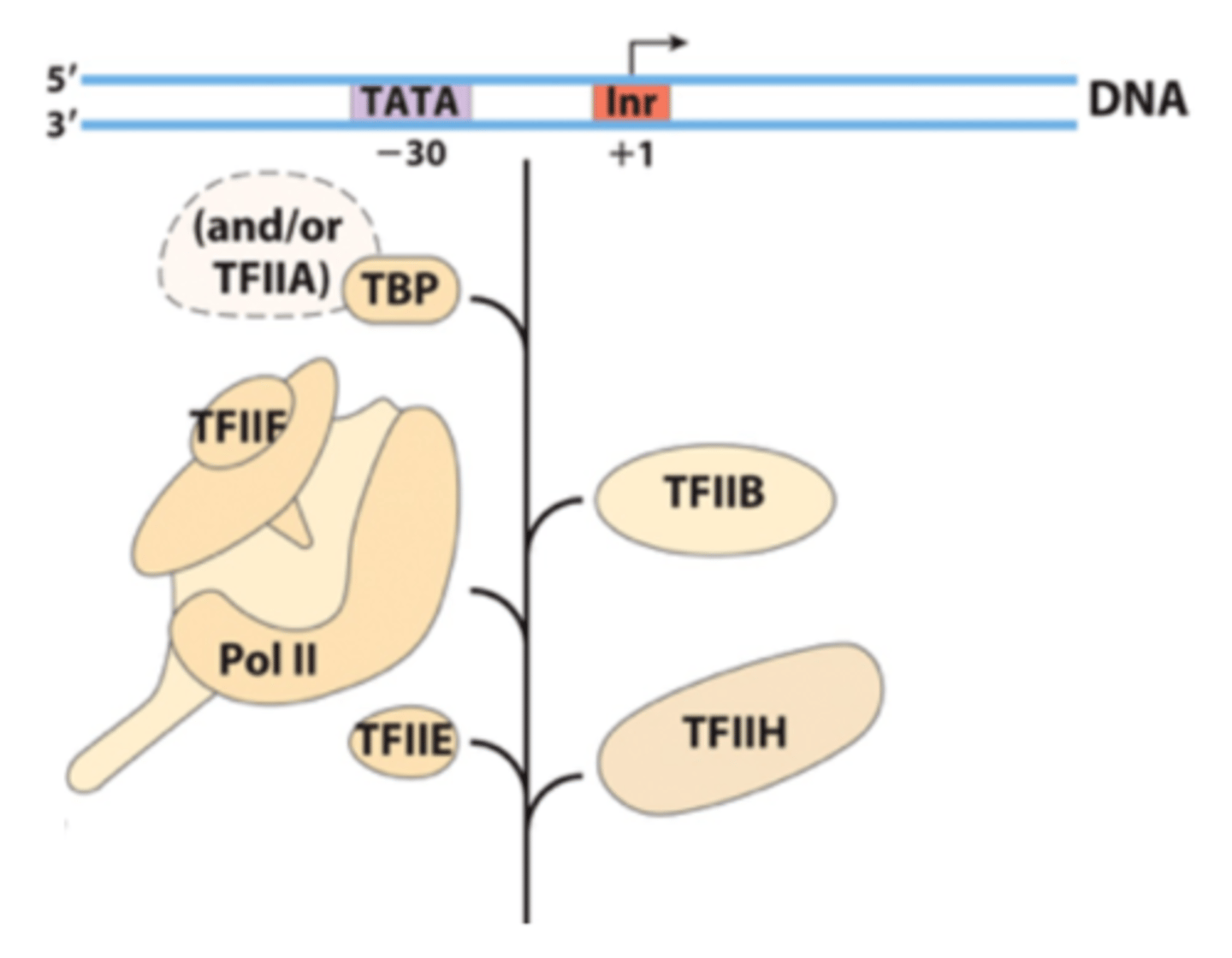
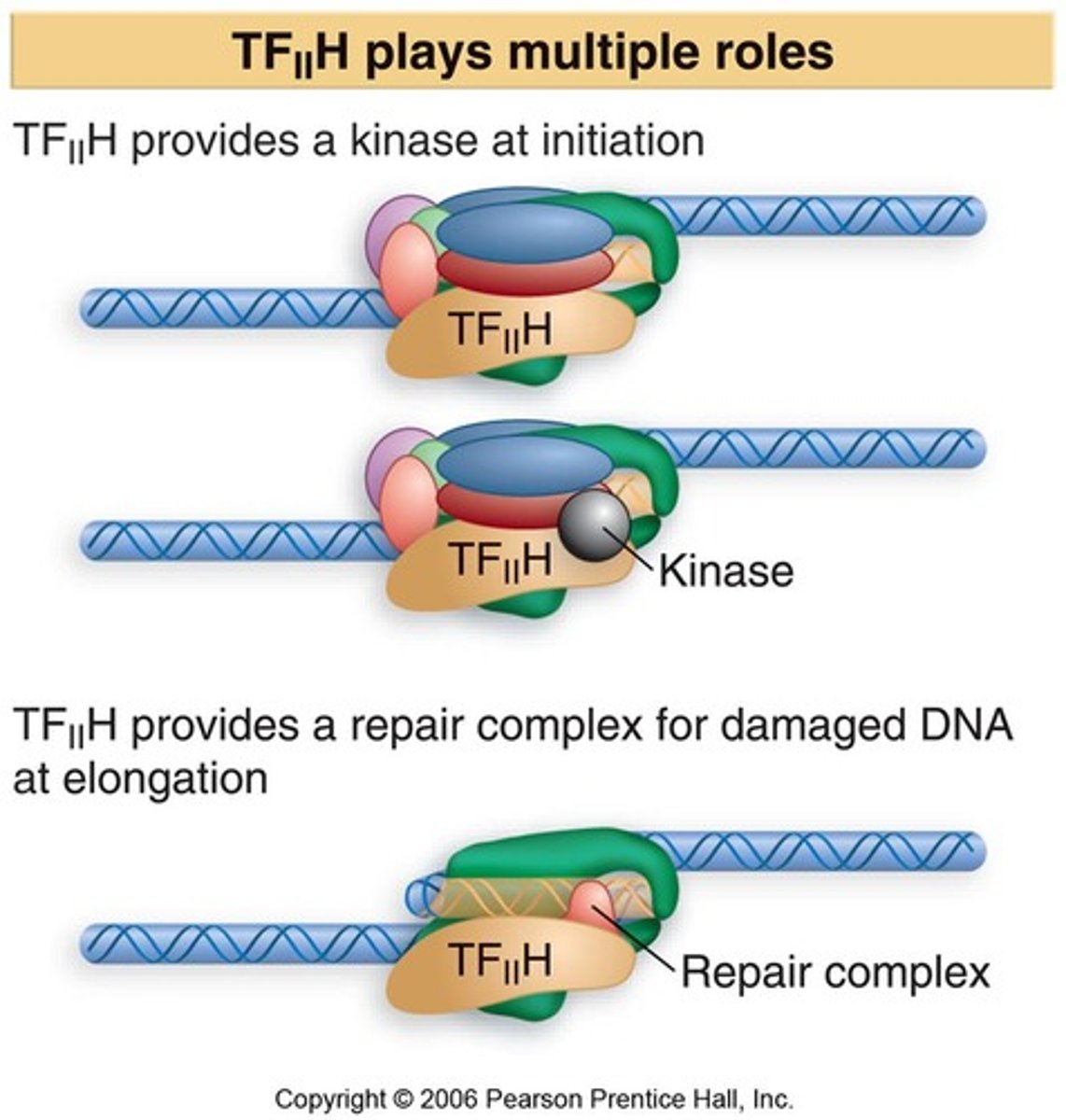
TFIIH
A multisubunit protein that has multiple roles.
- certain subunits act as helicases and promote the formation of the open complex.
- Other subunits phosphorylate the carboxyl terminal domain (CTD) of RNA polymerase II, which releases its interaction with TFIIB, to allow RNA polymerase II go through elongation
- has kinase activity
Mediator
a multisubunit complex that mediates the effects of regulatory transcription factors (that bind to enhancers or silencers) on the function of RNA polymerase II
----------------------
- affects the function of RNA poly. II
- can influence TFIIH to phosphorylate CTD
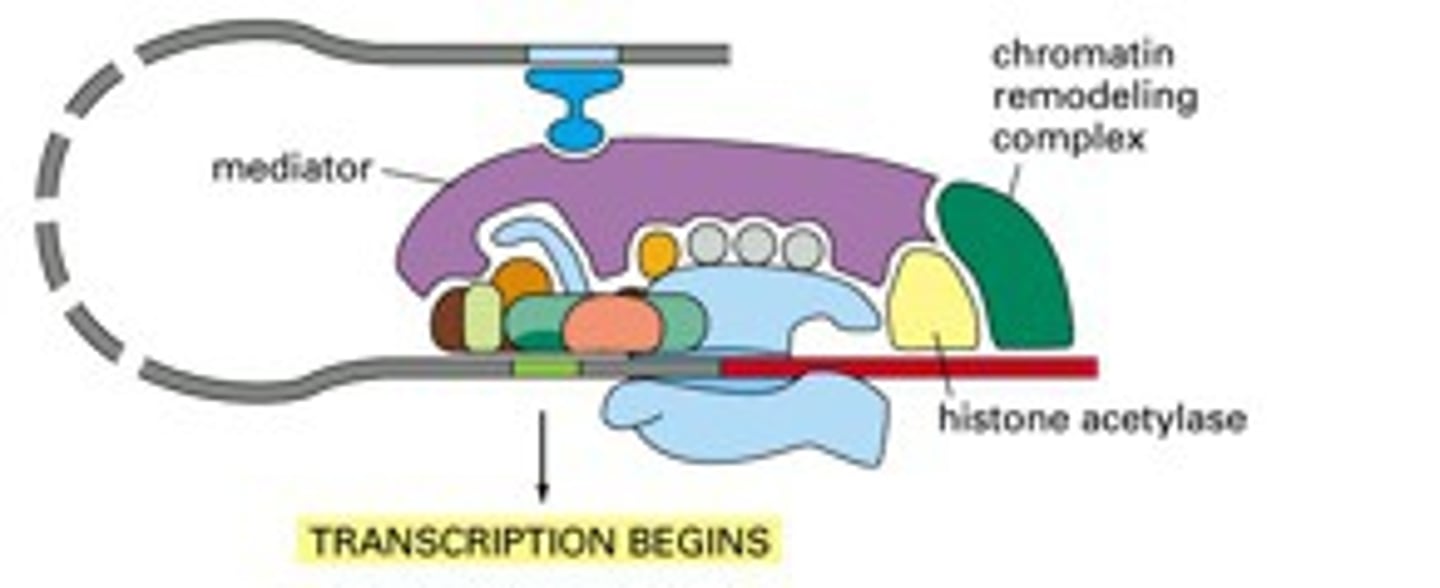
mediator abilities
can phosphorylate the C terminal domain of RNA pol. II which
- regulates TFIIH to phosphorylate to C terminal domain
----------------------
plays a big role in the switch between transcriptional initiation and transcriptional elongation
Eukaryotic RNA polymerases
- RNA pol I.
- RNA pol II.
- RNA pol III.
----------------------
all 3 are very stucturally similar and are composed of many subunits
RNA pol I (eukaryotes)
transcribes rRNA
- except for the 5s rRNA
RNA pol II (eukaryotes)
- transcribes all protein-encoding genes (mostly mRNA)
- transcribes some small nuclear RNA (snRNA) genes for splicing
RNA pol III (eukaryotes)
- transcribes tRNA genes , including 5S rRNA gene and micro RNA genes
eukaryotic genes main features
- core promoter
- regulatory elements
core promoter
- consists of the TATA box
- consists of the transcriptional start site
----------------------
its importance in determining the precise start point for transcription
basal transcription
core promoter can produce a low level of transcription on its own
Splicing
the process of removing introns and reconnecting exons in a pre-mRNA
- does not occur in prokaryotes (bacteria)
- can only occur in eukaryotes (within the nucleus not cytoplasm)
----------------------
possibilities include:
- exon 1 + 2 + 3
- exon 1 + 3
5' splice site
the 5' end of an intron where cleavage takes place in RNA splicing
branch point sequence
consenced section within a intron that contains adenine nucleotide (A)
- helps break down phosphodiester backbone
3' splice site
the 3' end of an intron where cleavage takes place in RNA splicing
Spliceosomes
active cutting complex that consist of small nuclear ribo-nucleo-proteins that recognize splice sites
----------------------
is a mechanism of RNA splicing
snRNPs
small mRNA and protein molecules that recognize the splice sites, join with additional proteins to form a spliceseome
----------------------
Types:
- U1
- U2
- U3
- U4
- U6
snRNP functions
- binds to intron sequences
- recognizes intron/exon boundaries
- hold pre-mRNA in the correct configuration
- can catalyze chemical reactions
- can convalently link exons
U1 snRNP
binds to 5' splice site
(is complementary to 5' splice site)
U2 snRNP
responsible for binding to the branch point sequence within the intron
U3 + U4 + U6 snRNP
snRNPs that combine with U1 and U2 to form a splicesome complex (active cutting complex)
lariet structure
a ring of intron segments that has been spliced out of a messenger ribonucleic acid molecule by enzymes
skipping and using different 3' splice sites
the transcription will still be the same as your still cutting a 5' splice site and a 3' splice site
Capping
7' methyl guanosine (G) convalently attaches at the 5' end
- is about 20-25 bases long
----------------------
Three enzymes of 5' capping:
- 5' RNA phosphatase
- guanylyl transferase
- methylase (methyltransferase)
5' RNA phosphatase
opposite of kinase as it removes phosphate group on 5' end (gama phosphate end = farthest right phosphate)
guanylyl transferase
links Guanosine mono phosphate (GMP) to RNA by using Guanosine triphosphate (GTP)
Methylase (methyltransferase)
enzyme that transfers methyl to base
(is a methyl doner)
Functions of capping
(a) Aids in mRNA exit from nucleus
----------------------
(b) Recognized by translation initiation factors
- recruits small ribosomes (40s)
----------------------
(c) is important in stability in mRNA)
----------------------
With out a cap transcription wont occur
When does capping occur?
occurs as the pre-mRNA is being synthesized by RNA poly.II
cap binding proteins (roles)
- aid in the movement of RNA into the cytoplasm
-happens in the early stages of translation
- aids in the splicing of introns
Poly-A Polymerase (PAP)
An enzyme that takes ATP and cuts/releases phosphoenol pyruvate
poly-A tail
a sequence of 50-250 adenine (A) nucleotides added onto the 3' end of a pre-mRNA molecule
- can be found as an export out of the nucleus to cytoplasm (via nuclear pores)
----------------------
- is found in bacteria (but does not have a cap structure)
- is not transfer RNA (tRNA) nor ribosomal RNA (rRNA)
poly A tail function (eukaryotes)
increases half live of mRNA (increase mRNA age)
- allows for translation to translate more efficiently
Poly A tail in translation
is not encoded in the gene sequence but is rather added enzymatically after the gene is completely transcribed
regulatory elements
short DNA sequences that affect the binding of RNA polymerase to the promoter
- binds to transcription factors to influence rate of transcription
----------------------
Types of R.E.
- enhancers
- scilencers
Enhancers
responsible for stimulating transcription
----------------------
- are usually found in the -50 to -100 region
scilencers
responsible for inhibiting transcription
----------------------
- are usually found in the -50 to -100 region
factors that control gene expression (location based)
- cis-acting elements
- trans-acting elements
cis-acting elements
DNA sequences that exert their effect only over a particular gene (within the same chromosomal strand)
- same strand
----------------------
ex: TATA box , enhancers , scilencers
trans-acting elements
regulatory proteins that bind to such DNA sequences
- must be guanine (G) that codes for transcription factor and bind to another promoter on a different chromosomal strand
- moves across strand
RNA Pol II transcriptional termination
Pre-mRNAs are modified near their 3' end with subsequent attachment of a string of adenines
-Transcription terminates 500 to 2000 nucleotides downstream from the polyA signal
-There are two models for termination
Models of termination
- allosteric model
- torpedo model
allosteric model of termination
after passing the polyA signal sequence, RNA pol II is destabilizes
(due to the release of elongation factors or the binding of termination factor)