Chapter 3: Digestion
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
56 Terms
Digestion
all intermediate processes, including absorption
absorption of food requires
conversion (= hydrolysis) of (non-soluble) polymers in (soluble) monomers
complex digestive tract with 4 zones:
reception zone (mouth)
transport zone (oesophagus)
digestive zone
mechanical and enzymatic pre-digestion (stomach)
complete digestion and absorption (small intestine)
absorption of water with formation and excretion of faeces
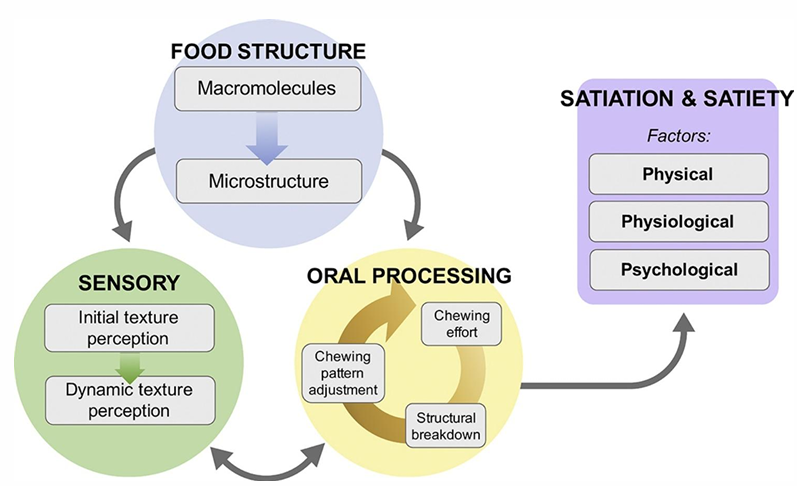
Reception zone - oral cavity
reduces particle size of food and stimulates enzymatic digestion by increasing the surface size
breakdown ~ intensity of chewing + residence time in the mouth
chewing = age dependent
A slower transformation of food structure induces …
satiation and satiety
summary oral processing
Formation of tooth caries
Tooth enamel
covered with a biofilm of
enzymes
glycoproteins
mucins
Bacteria in the mouth can adhere to the biofilm (e.g. Streptococcus)
Caries formation
Leuconostoc mesenteroïdes sectretes glucosyl-transferase which produces dextran starting from sucrose → dextran attaches to enamel and will attract acidifying bacteria → tooth caries
Treatment
oral hygiene
sugar replacers (e.g. fructose instead of sucrose)
Salivary glands
stimulated via taste- and smell receptors
stimulated by conditioned reflex
secretion is promoted by chewing
components saliva
ptyalin (alfa-amylase for hydrolysis of endogenous alfa-1,4 bonds in starch)
HCO3- = creating neutral environment for ptyalin
Cl- = activation of ptyalin
mucins = glycoproteins that glue and lubricate the food
lysozyme
IgA
papilla
taste buds on tongue that only react to soluble components
Tastes of tongue summary
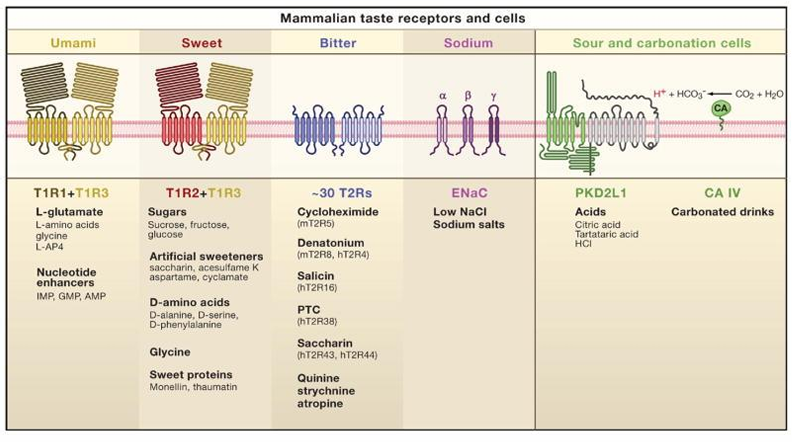
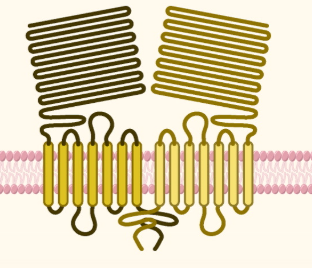
Tastes of tongue: Umami
Receptor
T1R1 + T1R3
Recognition
L-glutamate
Nucleotide enhancers: IMP (inosin) , GMP (guanosin)
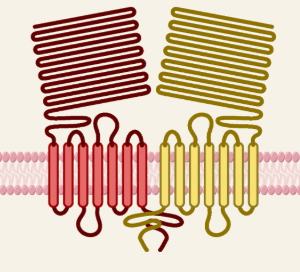
Tastes of tongue: Sweet
Receptor
T1R2 + T1R3
Recognition
Sugars (glucose, fructose, sucrose)
Sugar alcohols (sorbitol)
D- amino acids
artificial sweeteners (saccharin and aspartame)
proteins (monellin, brazzein)
Tastes of tongue: Bitter
Receptor
T2R receptor (very sensitive, but lack in selectivity
Recognition
salicin
phenyltiocarbamide (PTC)
6-n-propylthiouracil (PROP)
saccharin → in high concentrations
denatonium
Tastes of tongue: Salty
Receptor
ENaC = epithelial Na channel → not necessary in humans
Recognition
Na+
other ions (such as K+)
dipeptides
!! Narowness of alfa-, beta- or gamma-canals/loops => only sodium can pass through
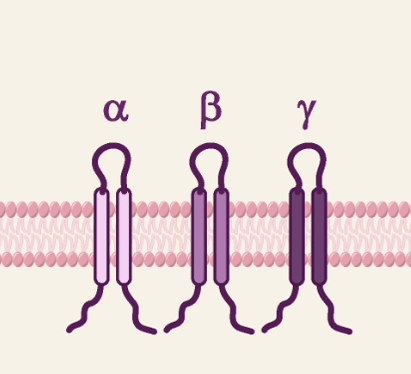
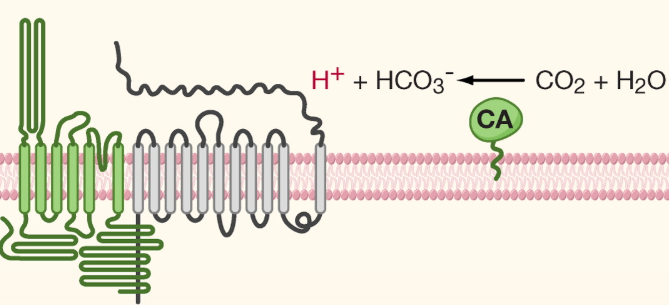
Tastes of tongue: Acid
Receptor
PKD2L1
Extra
CA = carbonic anhydrase: converts CO2 into bicarbonate ions and a proton (e.g. carbonated drinks)
Recognition
intracellular proton concentrations (pHi)
Extracellular protons are taken up by taste cells or organic acids = dissociate in cytoplasm
Taste also plays a role in
Food intake
Immune function
Glucose uptake
Transsit time
Insulin release
G protein coupled receptors (GPCR)
role in nutrient sensing including
sweet taste receptors
amino acid taste receptors
free fatty acid receptors
Gustary G-protein (gustducin)
linked to GPCR lead to IP3 mediated release of intracellular calcium and activation of cation channel
How taste developes
When mother during pregnancy or during lactation drinks or eats certain food, the aromatic components from the food are transferred to the fetus (during pregnancy) or the infant (during lactation) → influence of taste at young age
bitter suppression
suppressing bitter tastes: Aspartame > sodium salts
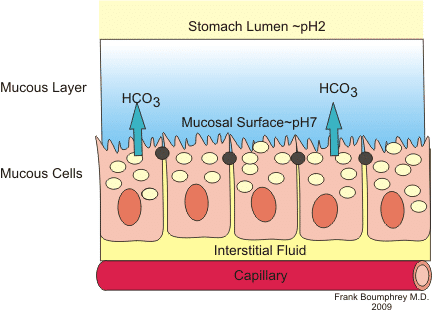
Stomach: mucus layer
protects wall of stomach against extreme environment in the lumen
secreted by mucous (goblet) cells
° mucine = glycoproteins containing cysteine and O-linked and N-linked oligosaccharides
low pH + protease activity
Defence and repair: antimicrobial proteins (lysozyme) + trefoil peptides (acid and enzyme resistant)
Stomach: epithelial wall
Mucosa / Mucous membrane
Large folds
smoothened when the stomach is filled with food
Foveolae/gastric pits
between folds
grooves with exits for the different glands
Mucous cells
produce mucine
Gastric glands
Chief cell/ zymogen cells
produce enzymes (pepsinogen + lipase)
Parietal cells
excrete HCl
use high amounts of energy
secrete the intrinsic factor that enables the absorption of vitamin B12 by the small intestine, otherwise B12 would be destroyed by stomach acid
ECL cells = Enterochromaffin-like cells
situated near parietal cells
aid in production of HCl by release of histamine
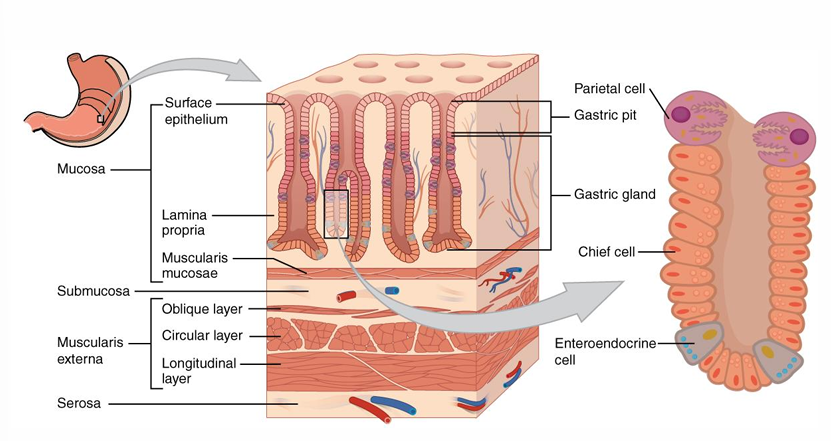
Summary secretion of each part stomach
Origin | Secrete |
Goblet cells/ mucous cells | mucines |
Gastric chief cells | pepsinogen and lipase |
Parietal cells | HCl and intrinsic factor (vitamin B12) |
Gastric enteroendocrine cells (pylorus) = Enteroendocrine G cells | gastrin |
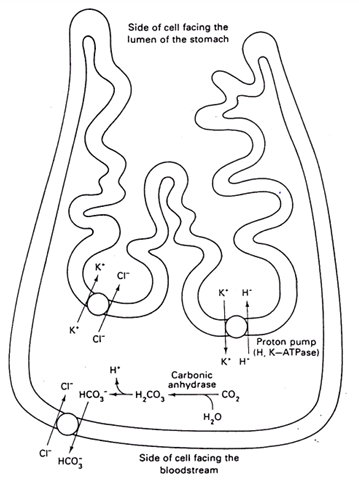
HCl production by parietal cells
1) A chloride-bicarbonate transporter in the basal membrane supplies Cl- which is released in cotransport with K+ by a KCl transporter at the apical membrane
2) H+ (produced by carbonic anhydrase) is released in the stomach lumen by an H+/K+- ATPase
Rules:
Na+ is always extracellular —> easy transport to intracellular
K+ is always intracellular —> easy transport to extracellular + can take Cl- with him
Gastrin
stimulates the movement of the corpus and release of hydrochloric acid
pylorus produced more gastrin when the wall is more stretched
Cell renewal in stomach
by displacement of non-differentiated cells of the corpus zone, which have high mitotic activity
cells | age | replaced by… |
mucus-producing cells | 4 days | non-differentiated cells |
mucosal neck cells, the zymogen head cells, wall cells | / | non-differentiated cells |
head and wall cells | few months | non-differentiated cells |
when small defects arise: non-differential cells originate again from differentiated cells and epithelium is covered in extra mucosal layer
fecal particle size depends on:
SCFA (= Short Chain Fatty Acids) production
residence time
microbiome diversity
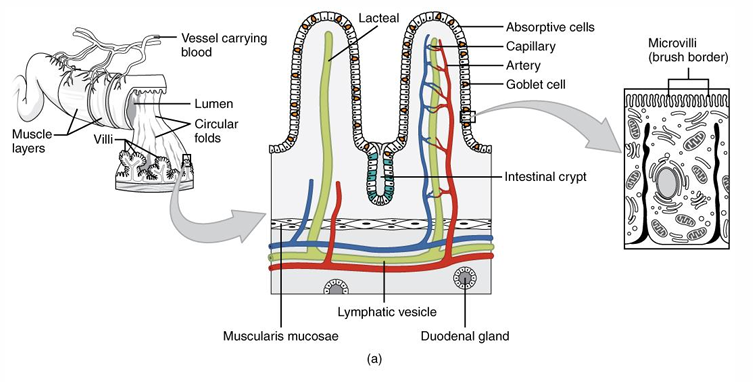
Small intestine
= site of enzymatic digestion
Wall composition (outside → inside)
serosa
muscularis externa
submucosa
inside mucosa
Crypts of Lieberkühn
Enlargen absorption surface
circular folds of Kerkring
villi
microvilli
duodenum => secretion
epithelial cells = glands of Brühner near Crypts of Lieberkühn → secrete intestinal juice
Liver → secretion of bile (concentrated by absorption of water and salts)
pancreas → secretion of enzymes (proteases, lipases, carbohydrases)
jejenum => absorption
reabsorption => ileum
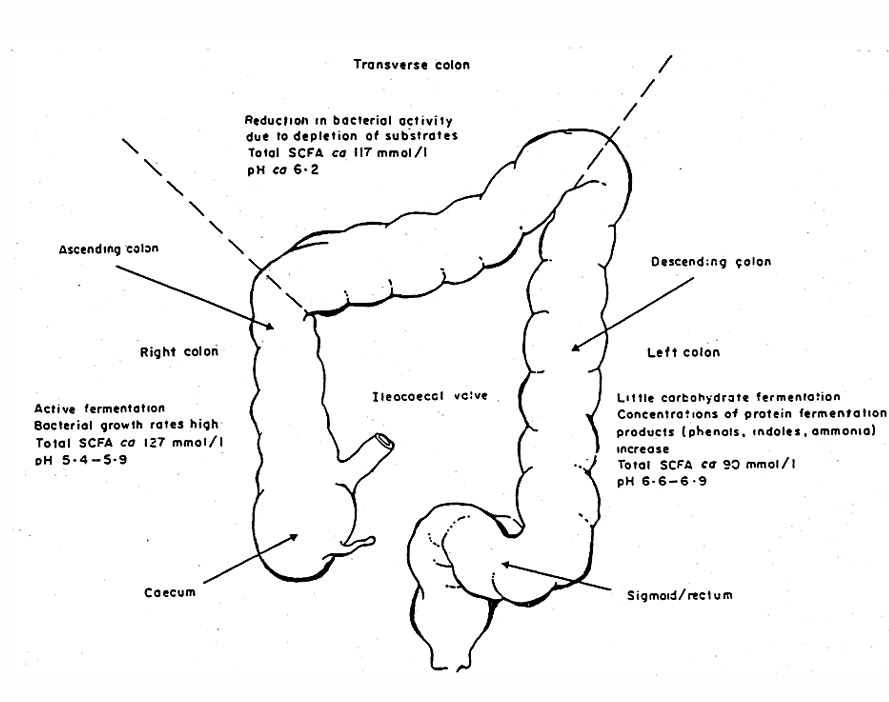
Large intestine
= site of microbial fermentation
no villi, but monolayeer of cylindrical apithelium consisting of mucusproducing cells and enterocytes with short microvilli
ascending
transverse
descending
Villi
absorbing cells
mucus secreting cells
layer of microvilli containing polysaccharide fibers = Glycocalyx
Why is it important that the mitotic activity of non-differentiated stem cells in the crypts is not disturbed?
lifespan of inestinal ephithelium villi = 2 to 3 days
Reduction of turnover (e.g. infection, bad nutrition) → leads to shorter villi and reduces absorption
people suffer from malabsorption of food because of a reduced intestinal surface
GALT (What? Composition? Function?)
= Gut Associated Lymphoid Tissue
immune system of the intestine
Composition
ephithelial M-cells
macrophages
dendritic cells (DC)
B- and T-lymphocytes
Function
Immuno-exclusion = preventing uptake of MO and toxins
Immuno-elimination = neutralization of antigens during infection
Immuno-regulation = making a distinction between nutrients and potentially hazardous antigen
Involved in
inflammatory bowel disease
colorectal carcinogenesis
GALT - small intestine vs large intestine
Small intestine | Large intestine |
|
|
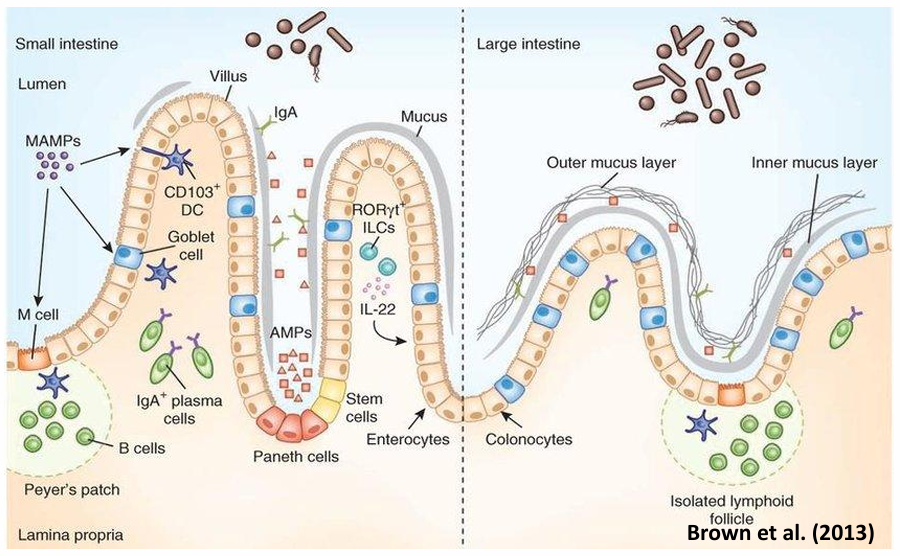
s-IgA
= secretorial immunoglobine-A
complex of IgA + secretory component
resistant against hydrolysis
secreted in intestinal lumen
Caco-2 cell
cultured in monolayer with microvilli + expression of small intestinal enzyme activities on the apical side
tight junctions between cells
semi-permeable filter support → more free access of ions and nutrients on both apical and basal sides of the monolayer
growth intestine
= defense mechanism against colon cancer
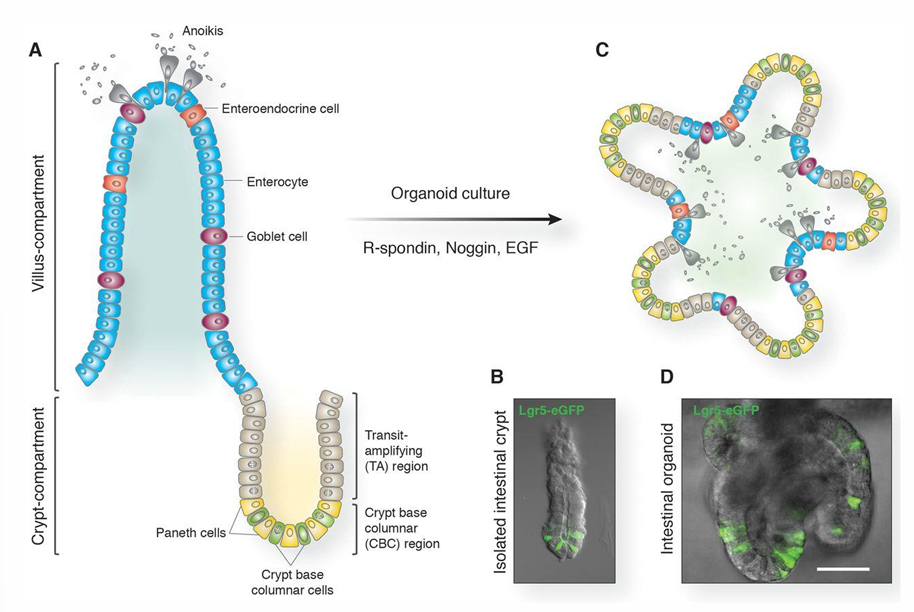
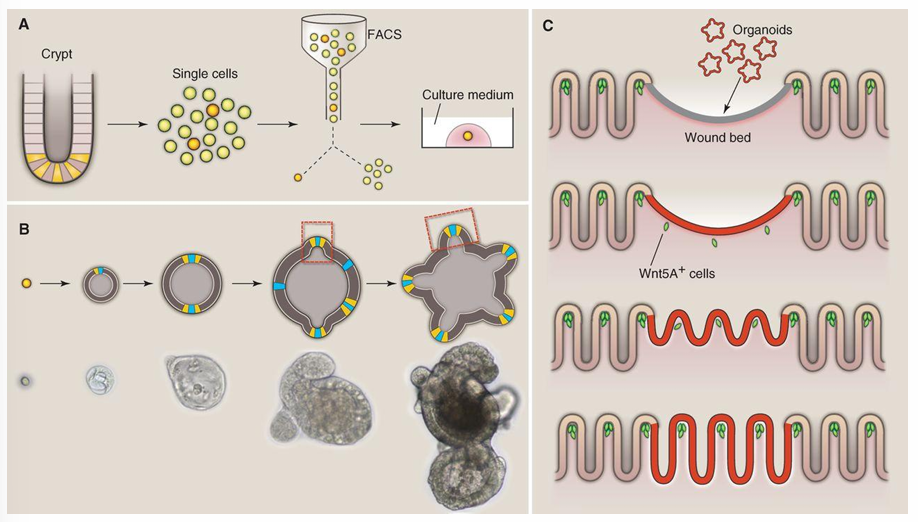
paneth cells stay in the crypt base with stem cells
1) stem cells produce organoids (= self-renewing intestinal ephithelia = Transit-amplifying cells)
2) TA cells proliferate rapidly and move up the walls of the crypt
2) As cells move upwards they begin to differentiate into goblet cells and enterocytes
3) Differentiated cells reach the villa → apoptosis → cells are shed into the lumen of the small intestine
Migration → cell death: 3 - 4 days
Movements stomach
During eating
Peristaltis = alternating contraction and expansion of circular and longitudinal muscle layer
Segmentation = rhythmic contraction of circular muscle layer
Between meals
Interdigestive migrating motor complex (MMC) = makes sure that all remaining food in the stomach and small intestine is transferred to the large intestine → interrupted after the intake of a meal
Trajectory food from esophagus to stomach (from discontinuous eating → continuous digestion)
1) upper sphincter relaxes + epiglottis prevents access to trachea
2) peristaltic movement to the stomach
3) Entering the stomach
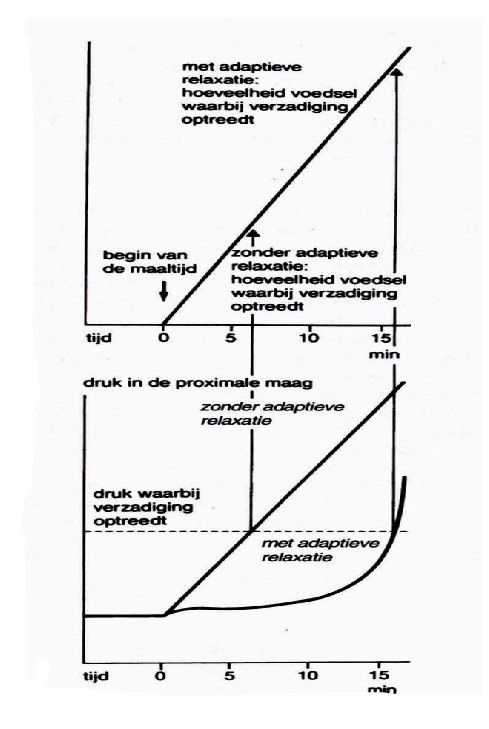
adaptive relaxation: corpus and fundus relax = prevents the pressure in the stomach to become too high and the feeling of satiation to occur too fast when the stomach contents during meal increases
postprandial phase: peristaltic wave in the antrum + closure of the pylorus → food fragments are mixed with gastric juice
Smaller particles can pass and arrive gradually in the duodenum
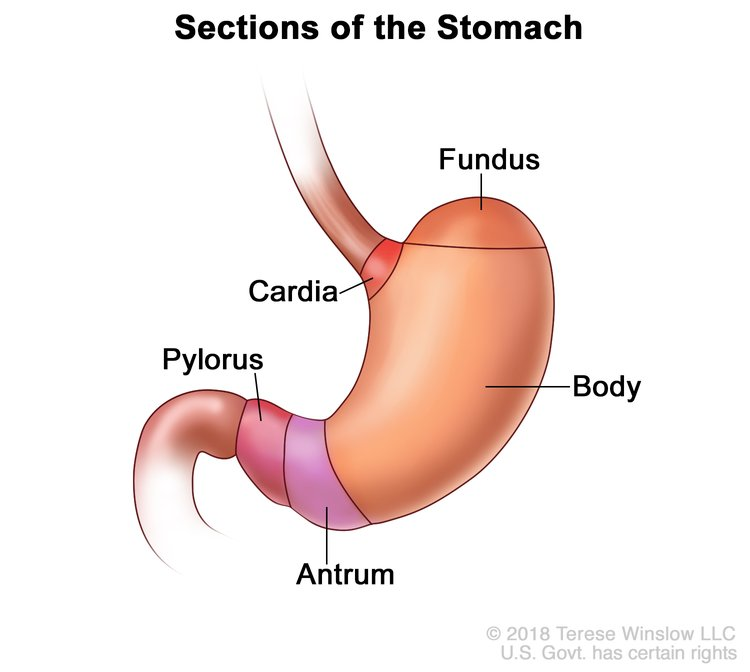
Cardia: welcomes the food coming from the oesophagus via peristaltic movements
Fundus + Corpus: relax when food enters the stomach to prevent too high pressure and early satiation (adaptive relaxation)
Antrum: peristaltic movements around the food and release of gastric juices
pylorus: transition from stomach to intestine
Ileal brake
= When not absorbed fat reaches the ileum → reflex inhibits further emptying of the stomach
Gastro-colic reflex
increasing contractions in the colon when food enters the stomach to make place for new food
Tracers
= easily identifiable components that behave in the same way as the examined material and do not noticeably influence the characteristics of the unit under examination
isotope method
13C-tracer is absorbed in the small intestine after which it participates in respiratory metabolism to produce 13C-CO2 which is excreted in expired air
in solid and liquid meal
Endogenous hydrolic digestion (purpose, regulation, enzymes, barriers)
= hydrolysis by enzymes
Purpose
high molecular compounds → low molecular compounds (water soluble)
Regulation
pH (acid predigestion vs alkaline digestion) → activation of inactive form of the enzyme prevents autolysis
ions
Temperature
Main enzymes
proteases/peptidases
carbohydrases
lipases
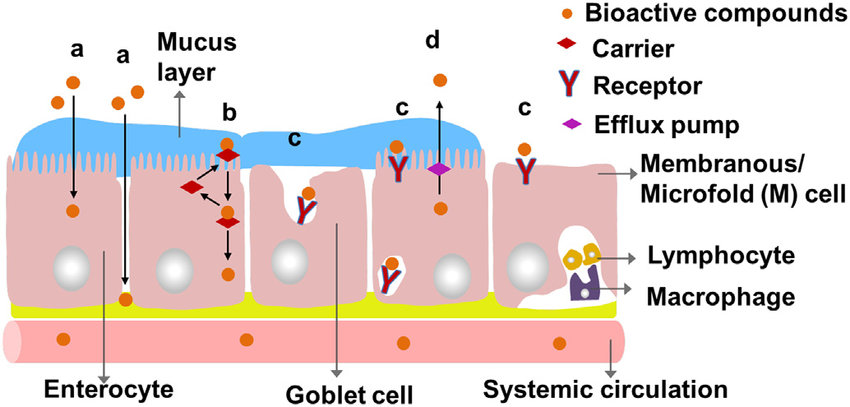
Barriers that need to be passed
1) The mucuslayer: diffusion of hydrophilic components
2) The apical membrane of the enterocyte: hydrophobic
3) The enterocyte
4) The basal membrane of the enterocyte: hydrophobic
Proteases/ peptidases
Location
stomach
small intestine
Autolysis
to avoid autolysis proteases are secreted as inactive zymogens which are activated in the lumen
Endopeptidases
aromatic AA
pepsin
chymotrypsin
basic AA
trypsin
inactive zymogen | location activation | protease |
pepsinogen | stomach | pepsin |
trypsinogen chymotrypsinogen | pancreas, by: enterokinase trypsin | trypsin chymotrypsin |
Exopeptidases → clave AA from C- or N-terminal of oligopeptides
NH2 (glycocalyx)
COOH (pancreas)
inactive zymogen | location activation | protease |
procarboxypeptidases | intestinal lumen | carboxypeptidase |
aminopeptidases | intestinal epithelium | aminopeptidases + dipeptidases |
Transport of dipeptides to bloodstream
1) absorbed in the enterocyte by non-selective peptide transporters (PEPT1 and PEPT2 = symporters of H+)
2) Hydrolysed by intracellular peptidases
3) Active uptake to the bloodstream by the influx of Na+ (energy provided by ATP)
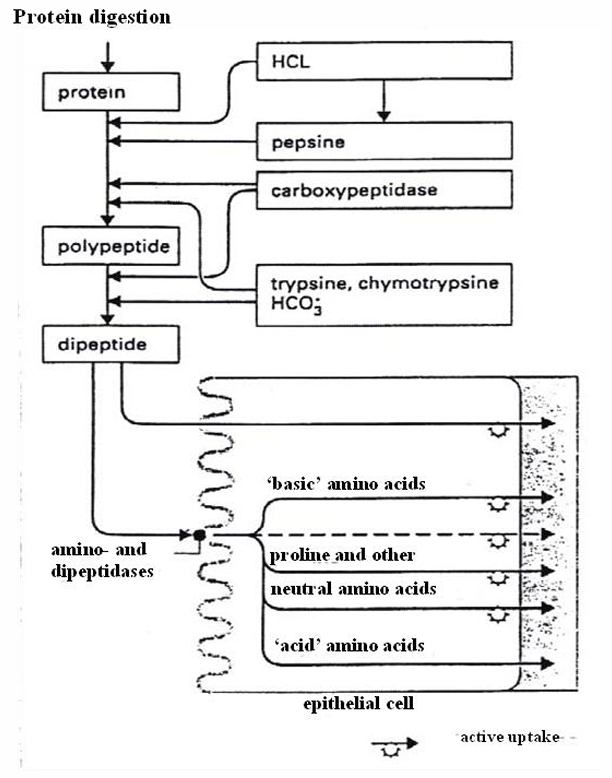
pinocytosis/ paracellular transport
during periods of specific needs some larger peptides and even proteins may enter the bloodstream intact
(e.g. pregnancy, lactation, infancy)
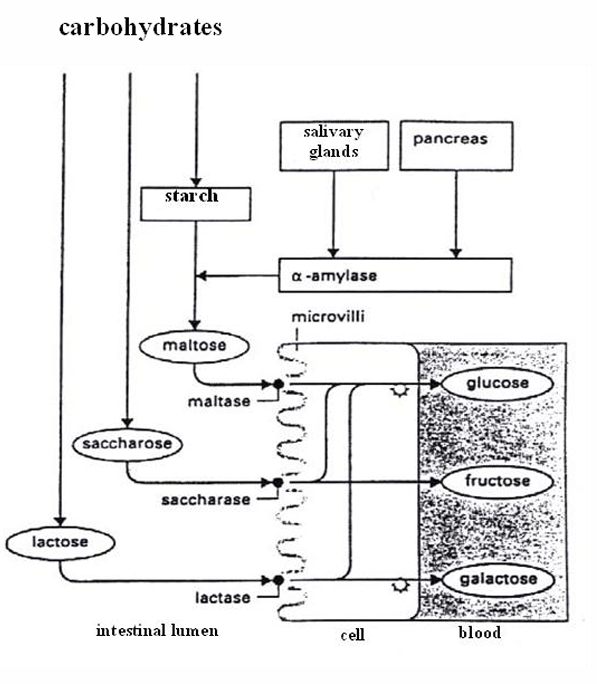
Carbohydrases
Amylase
starch: amylose (linear) + amylopectin (branched) → dextrin → maltose
alpha (animal) vs beta (plant)
require Cl- and light alkaline environment
saliva = ptyalin (alfa-amylase): hydrolyses alfa-1,4 bonds in starch
Glycosidases
Maltase/sucrase(invertase)/lactase
= extracellular glycosidases
dissacharides → monosacharides (glucose, fructose, galactose)
Isomaltase
adhered to sucrase → sucrase-isomaltase
hydrolyses alfa-1,6 bounds in starch (branches)
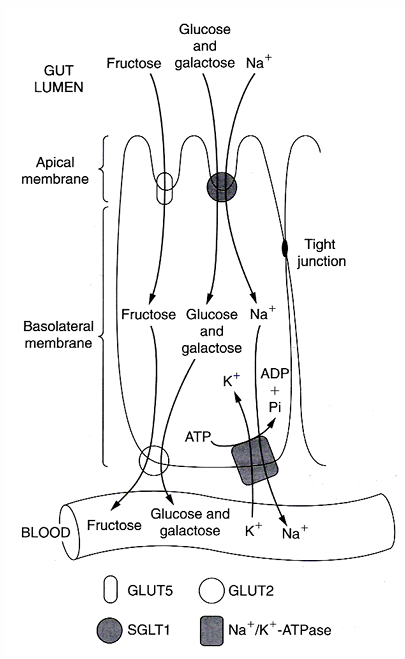
Transport to enterocyte
glucose/ galactose: SGLT1 (= sodium-glucose transport protein-1) + parallel influx of Na+ with energy use
fructose: GLUT5 by facilitated diffusion → only a proportion of fructose can be absorbed + the remaining stays in the lumen causing osmotic diarrhea or is changed to lipids (= liver fattening
Transport to blood
Glu,gal and fru use the same transporter: GLUT2
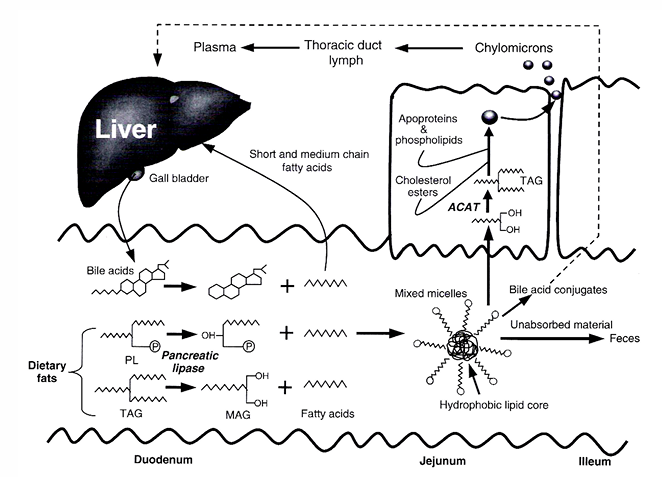
Lipases
Characteristics
less substrate specificity
primary alcohol groups → beta or 2-monoglyceride and FFA
lipolysis occurs in stomach
pancreatic enzyme secreted in the duodenum
Steps
1) pancreatic lipase:
bile salts: emulsify fat
Ca and co-lipase: break down bile salts + hydrolyse triglycerides
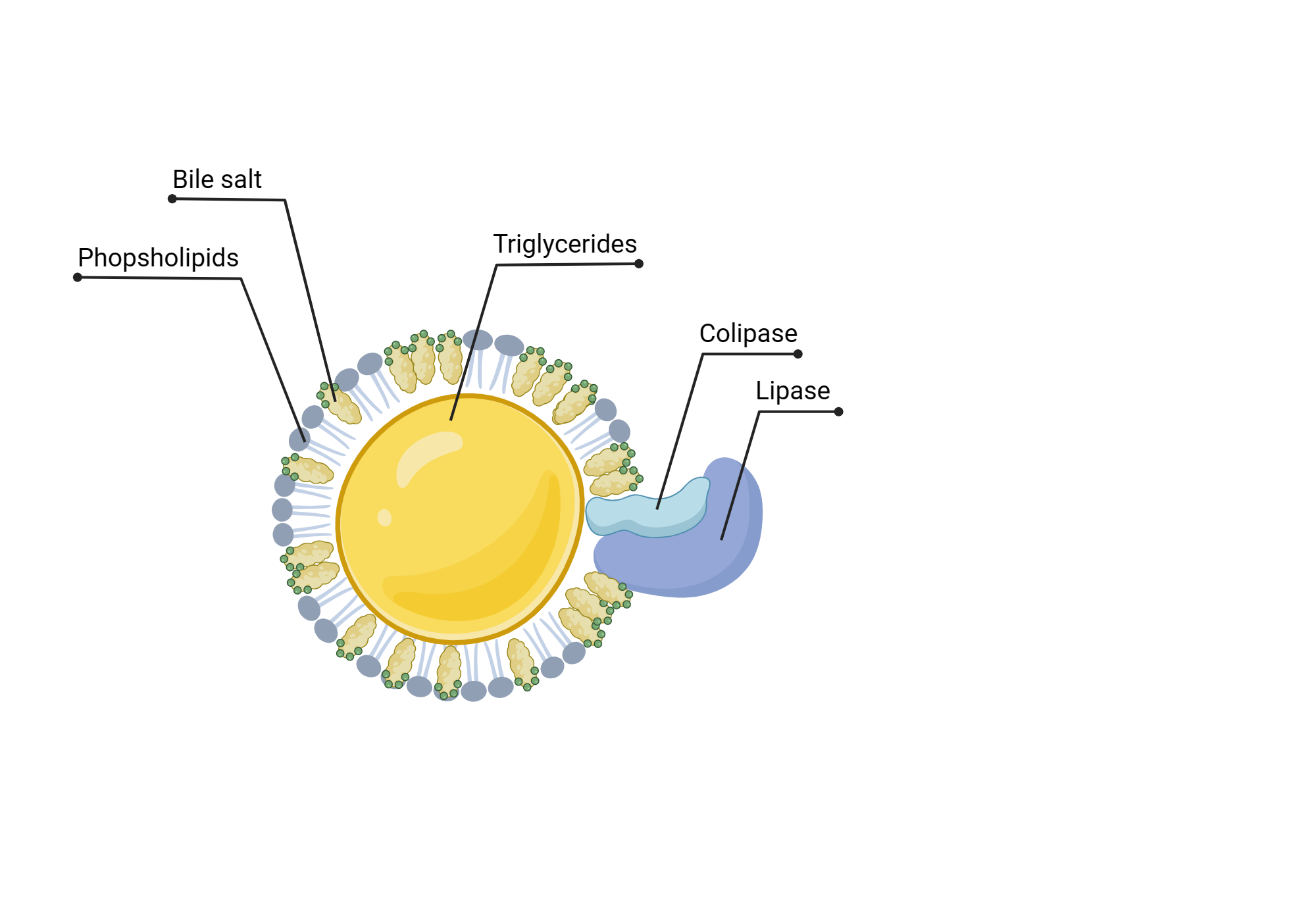
2)
triglycerides → fatty acids → micelle formation → monoglycerides and FFA’s are absorbed
bile salts are absorbed in ileum → transport via portal vein → liver → reused in gall-bladder
3) in enterocyte
FFA’s and monoglycerides → co-enzyme A esters of FFA (requires energy) → triglycerides
triglycerides packed in chylomicrons (= lipoproteins) → tranpsorted to lymphatic system → taken up in blood stream
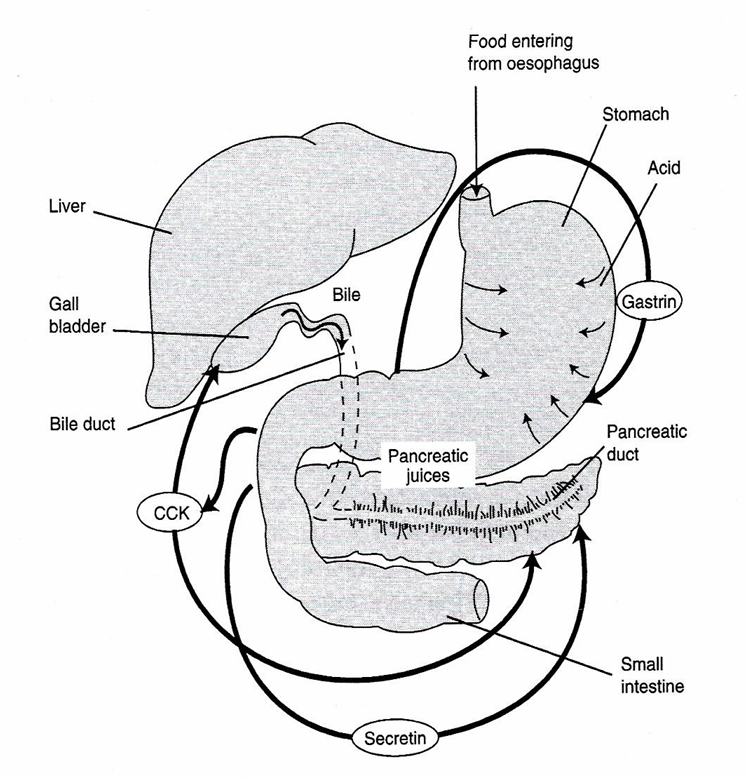
Gastro-intestinal peptides / hormones
Hormone | Target organ | Action | Stimulus |
Gastrin | Stomach | Secretion HCl Motility ↑ | Filling of the stomach |
CCK-PZ | Gall bladder, pancreas | Contraction, secretion pancreatic juice/ bile salts | FA and AA in duodenum |
Secretin | Pancreas, stomach | NaHCO3 ↑ stomach motility↓ | Food in stomach and intestine |
GIP | stomach, intestine | Intestinal juice ↑, motility ↓ | CH and fat in duodenum |
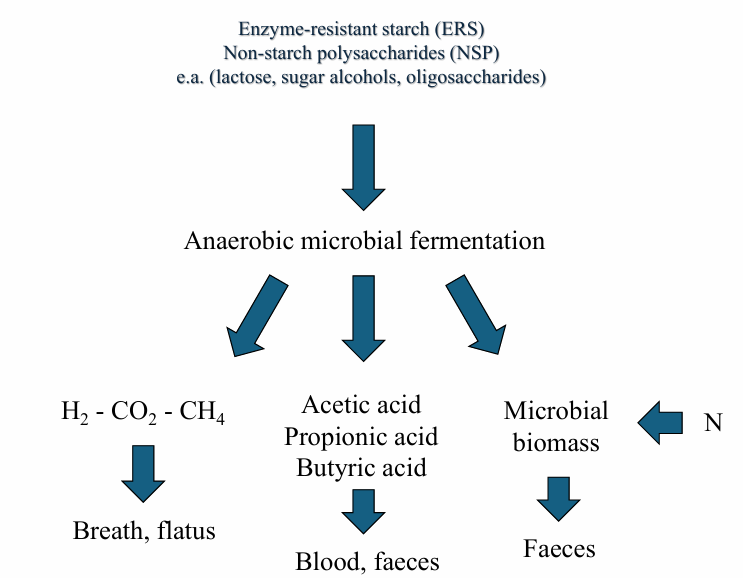
Humans/ MO interaction: competitive/fermentative model of digestion
1) humans digest the food according to competitive model
2) residues are fermented by MO in storage organ after stomach and small intestine (= colon): use food substances and nitrogen sources in anaerobic way
composition microflora: how is it determined?
1) The host
specificity adhesion and absorption capacity
rate of secretion and composition
motility in the intestine, frequency and amount of feed uptake
2) The food
form
composition
contamination
3) The MO
specificity adhesion → determines which MO can succesfully colonize the colon
growth factors
enzymes
Types of MO
1) Useful bacteria
Lactobacilli
Streptococci
Enterobacteriaceae
Bacteroïdes
2) Pathogens
Microbial pathways in the colon: carbohydrate metabolism
What: food polysaccharides
ERS = enzyme-resistant starch (alfa-bound hexose)
NSP = non-starch polysaccharides (beta-bound hexose)
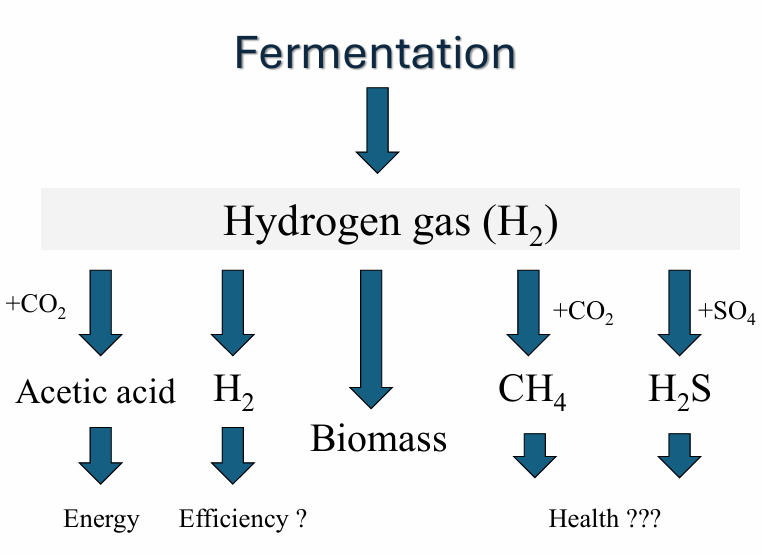
reductive acetogenesis: H2 → acetic acid
sulfate reduction: H2 → H2S
Microbial pathways in the colon: N-metabolism and microbial growth
protein → non-protein compounds (peptides, AA and NH3) → N-sources for microbial growth
Fermentative activity in the colon (order)
1) carbohydrates are broken down
2) protein fermentation starts to dominate when carbohydrate reserves are depleted → protein breakdown products (phenols, amines, ammonia) → negative for colon health (cancer)
Solution: remove tumor part in wall and replace by stem cells → redesign the wall
Characteristics of axenic/gnotobiotic animals
General
Axenic animals
improved turnover of feed and reduced peristaltis
alterations in morphology and contents intestine
no lactic acid and lactase
higher losses of urea and endogenous N
no branched and less saturated FA in faeces
Only conjugated primary bile acids in faeces
Possible deficit of vitamins B and K
More efficient intestinal absorption (antibiotics)!
Gnotobiotic animals
microflora is completely identified according to the accepted standards
Probiotics
= live MO that, when administered in sufficient quantities, confer a health benefit on the host
Examples
Bifidus
Lactobacillus
Faecalibacterium prausnitzii
Akkermansia muciniphila