BIO230 L4 eukaryotic gene regulation
1/35
Earn XP
Description and Tags
just the names of things
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
36 Terms
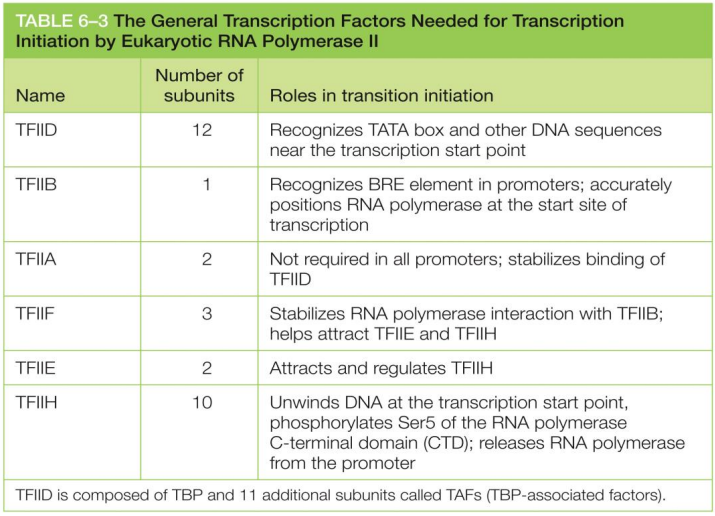
TFIID
general transcription factor
recognizes TATA box and other DNA sequences (other consensus sequences) near the transcription start point for initiation
bends DNA to highlight it for RNA polymerase recognition
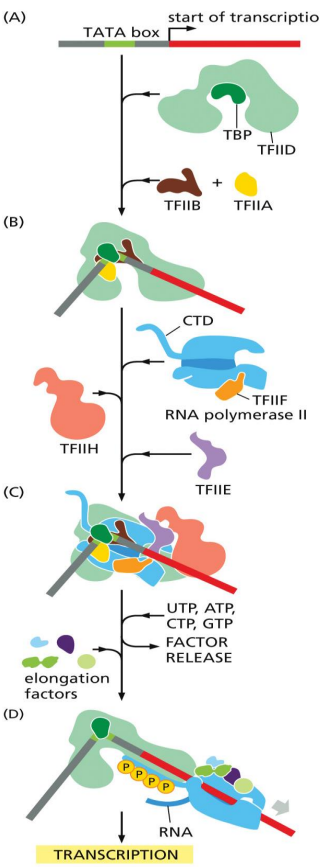
TFIIH
general transcription factor
unwinds DNA at the transcription start point
phosphorylates Ser5 of the RNA polymerase C-terminal domain (CTD)
allowing the recruitment and release of various protein factors required for mRNA processing
releases RNA polymerase from the promoter to initiate transcription
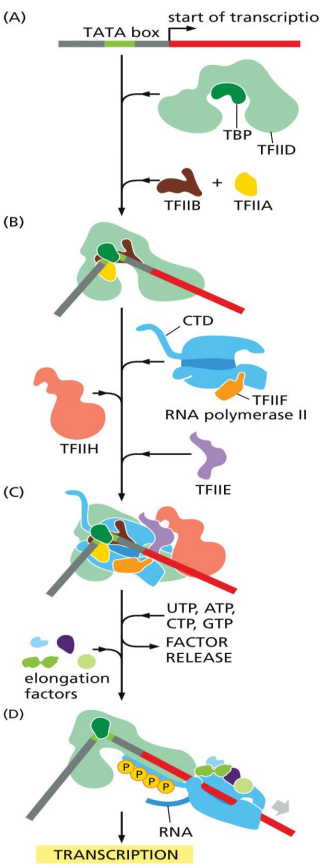
RNA polymerase II
transcribes protein coding genes in eukaryotes
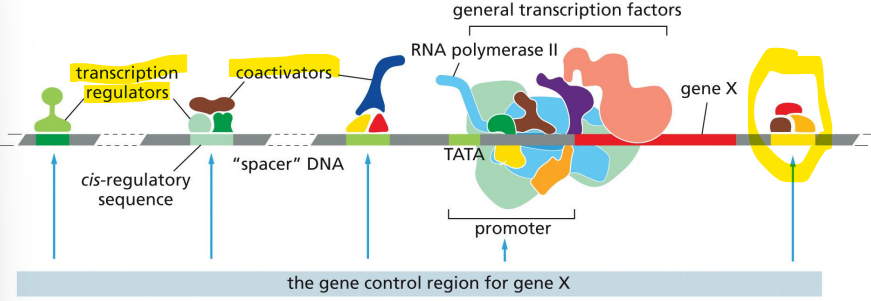
general transcription factors
help position RNA polymerase at promoters
TFIID, TFIIB, TFIIF, TFIIE, TFIIH for eukaryotes
sigma factor for prokaryotes
difference between eukaryotic genomes and prokaryotic genomes
eukaryotic genomes
lack operons
1 gene, 1 RNA molecule, 1 protein
packaged into chromatin (additional mode of regulation)
prokaryotic genomes
operons
multiple genes encoded on 1 RNA molecule
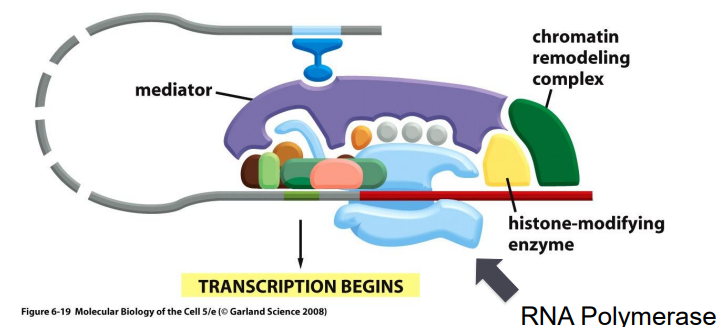
mediator
acts as an intermediate between many regulatory proteins and RNA polymerase in eukaryotes
regulatory proteins for eukaryotic gene expression
both activators and repressors
can act over very large distances, sometimes >10,000 bp away by DNA looping
often function as protein complexes on DNA
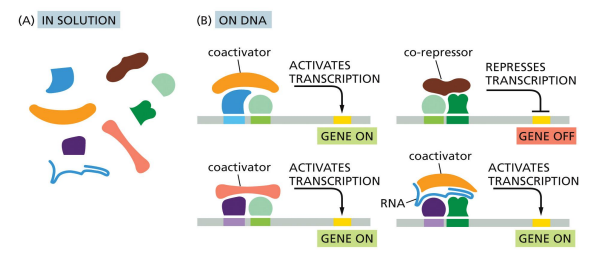
includes co-activators and co-repressors that do not directly bind DNA
coactivators and corepressors
only bind to proteins that are already bound to DNA, do not directly bind to DNA
works in a complex and aids activators and repressors with regulating gene expression
DNA binding domain (DBD)
a module of eukaryotic activator protein that recognizes specific DNA sequence
very specific about binding
binds to activation domain (AD) to adjust frequency/rate of transcription
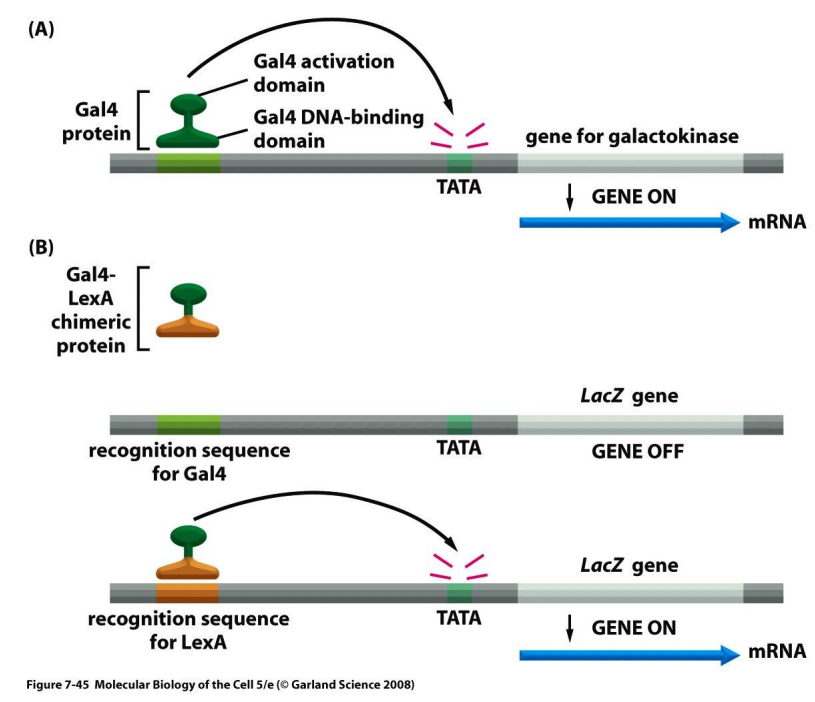
activation domain (AD)
a module of eukaryotic activator protein that accelerates frequency/rate of transcription
tends to be general about binding
binds to DNA binding domain (DBD) to recognize specific DNA sequence
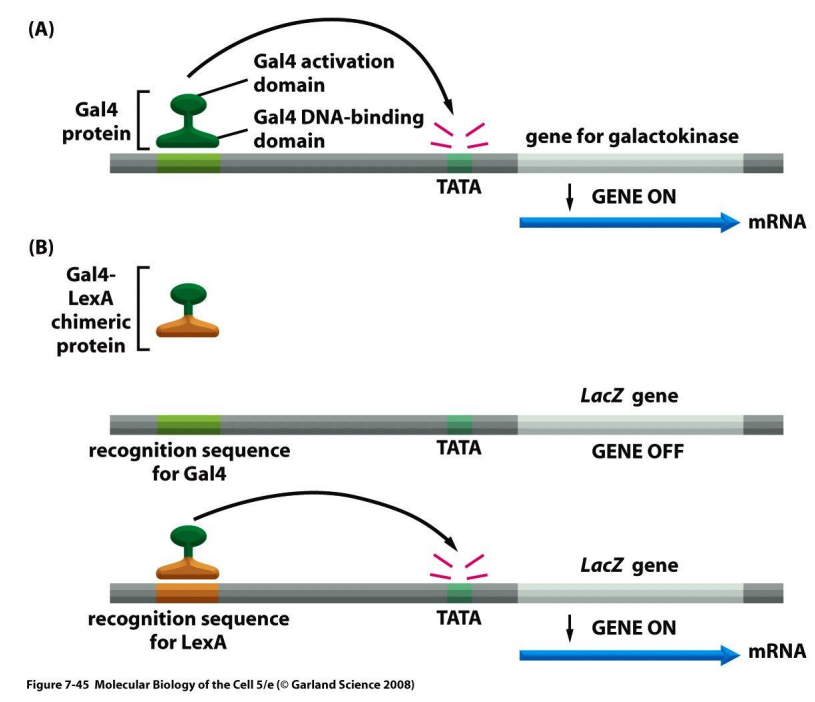
how activator proteins activate transcription
attract, position, and/or modify
general transcription factors, mediator, RNA polymerase II
by either
directly interacting with them
indirectly modifying chromatin structure
how activator proteins directly activate transcription
bind directly to transcriptional machinery or the mediator and attract them to promoters (like prokaryotic activators)
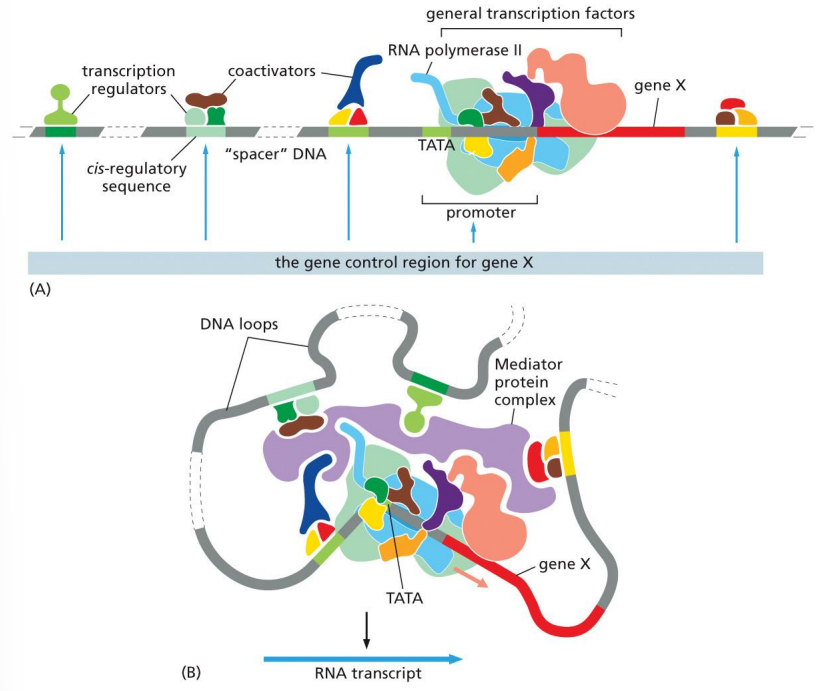
how activator proteins indirectly activate transcription
alter chromatin structure and increase promoter accessibility in 4 ways
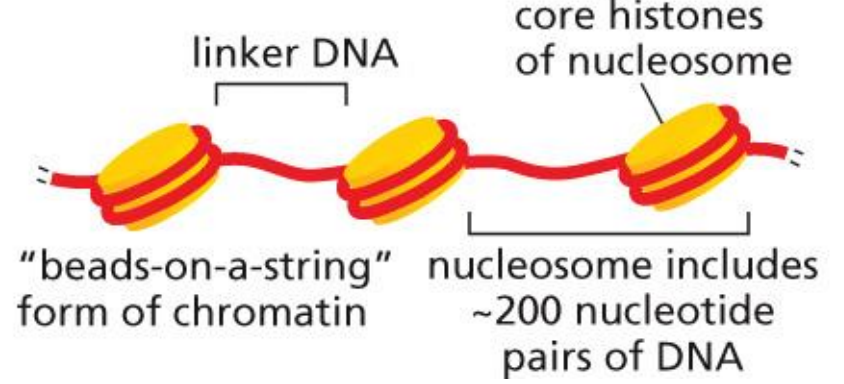
nucleosomes
the basic structure of eukaryotic chromatin
DNA wound around a histone octamer with 2 sets of H2A, H2B, H3, H4 creates a compact chromatin fibre
can be unwound or altered to increase promoter accessibility
histone tails are modified over the course of a cell’s life to meet its particular gene expression needs
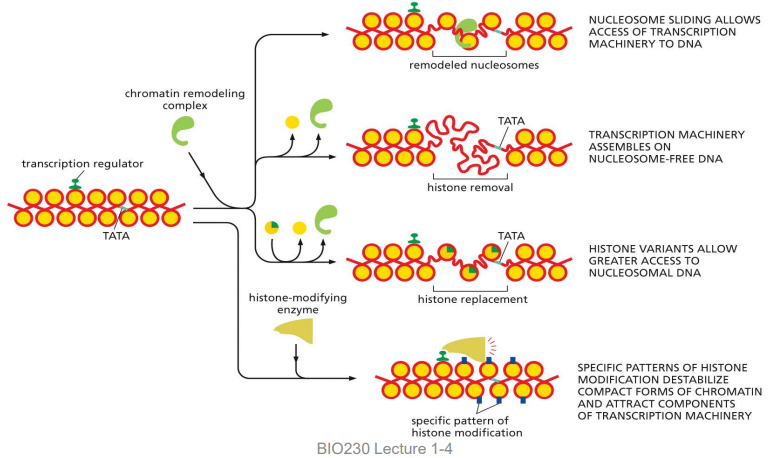
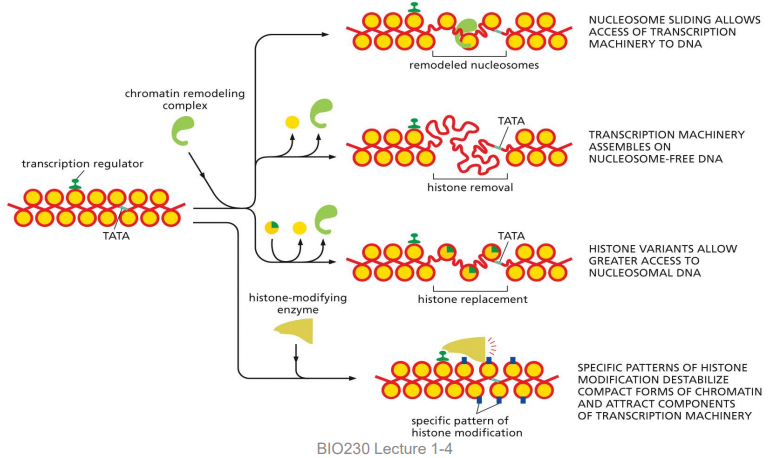
4 major ways activator proteins can alter chromatin
using chromatin remodeling complex:
nucleosome sliding
histone removal
histone replacement
histone-modifying enzyme:
specific pattern of histone modification
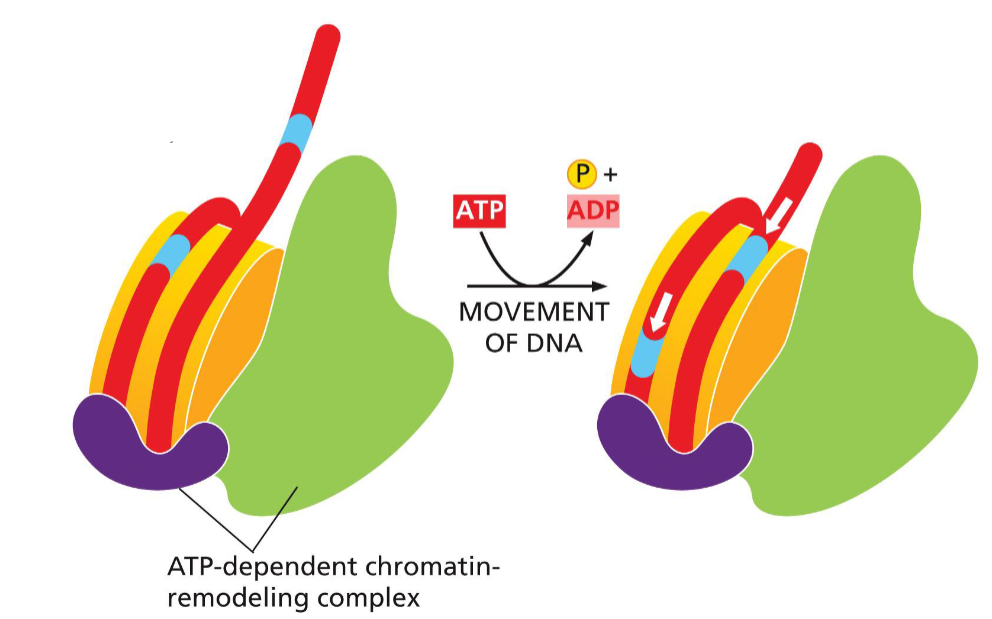
nucleosome sliding
nucleosome structure altered by ATP-dependent chromatin remodeling complexes to increase promoter accessibility
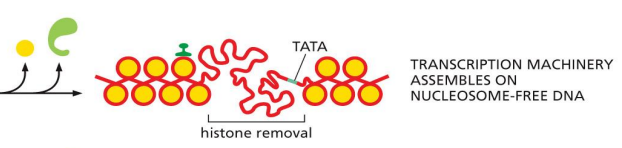
nucleosome removal
requires cooperation with histone chaperones
ATP-dependent chromatin-remodeling complex removes a histone core interfering with accessibility of target gene
ATP-dependent chromatin remodeling complex dissociates
transcription machinery assembles on nucleosome-free DNA
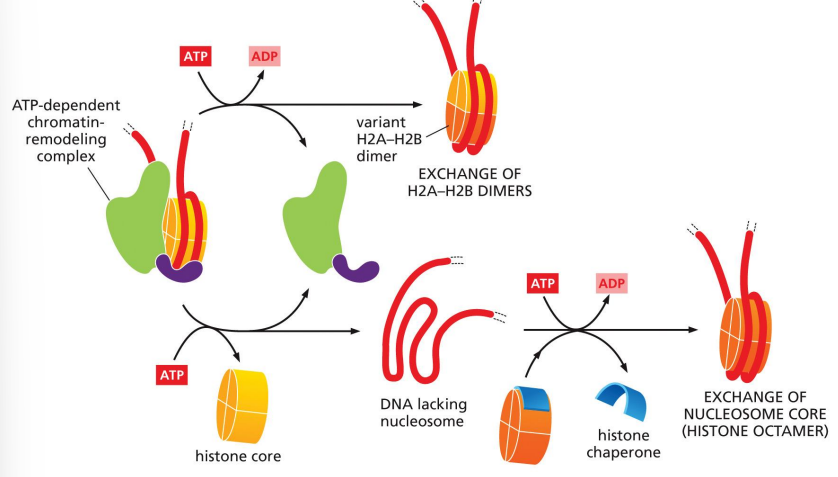
histone exchange/replacement
requires cooperation with histone chaperones
exchange:
ATP-dependent chromatin-remodeling complex creates a variant of histone dimer(s)
replacement:
ATP-dependent chromatin remodeling complex removes histone core, then histone chaperone exchanges for a new nucleosome core
histone variants allow greater access to nucleosomal DNA
chromatin remodeling complex
an ATP-dependent protein that modifies chromatin to regulate transcription
attracted to chromatin by transcription regulator
3 main functions:
nucleosome sliding
histone removal
histone replacement
histone modifying enzymes
AKA “writers”
produce specific patterns of histone modifications on specific amino acids of histone tails
patterns of histone modifications can increase/decrease gene accessibility
can destabilize compact forms of chromatin
can attract components of transcription machinery

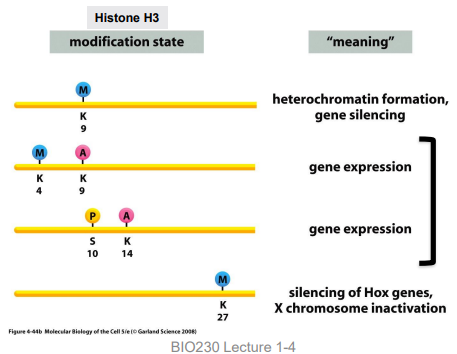
types of modifications in histone code
addition of phosphate group (phosphorylation)
performed by kinase enzyme
addition of acetyl group (acetylation)
performed by acetyltransferase enzyme
addition of methyl group (methylation)
performed by methyltransferase enzyme
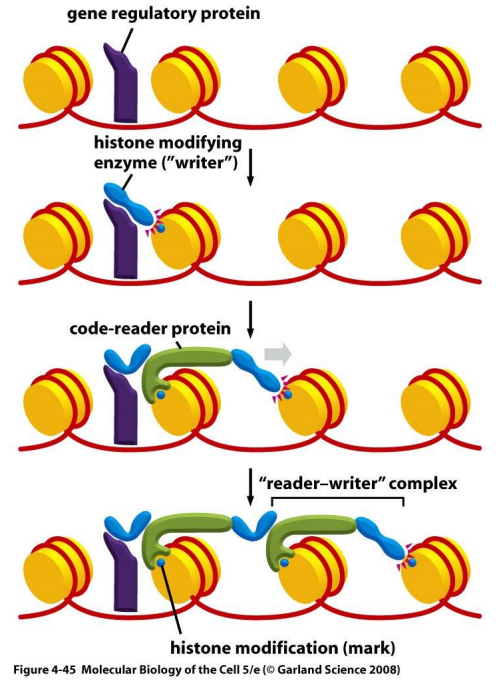
histone code
specific modifications to histone tails by histone modifying enzymes nicknamed “writers”
code “reader” proteins can recognize specific modifications and provide meaning to the code
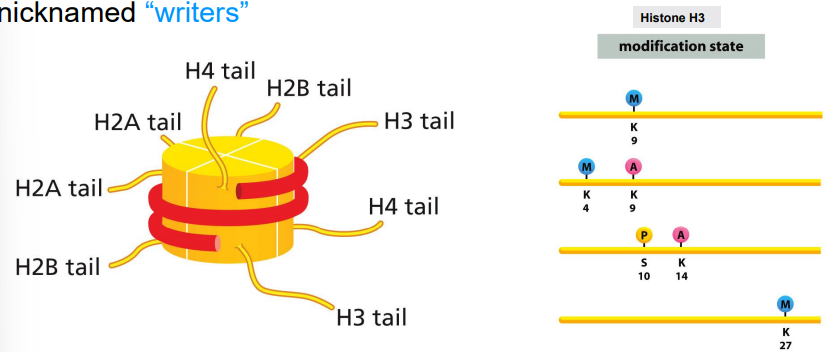
writer proteins
histone modifying enzymes that specifically modify histone tails for transcription regulation
part of the histone code
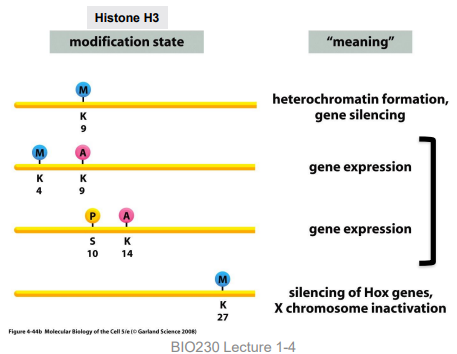
reader proteins
histone modifying enzymes that recognize specific modifications and provide meaning to the code
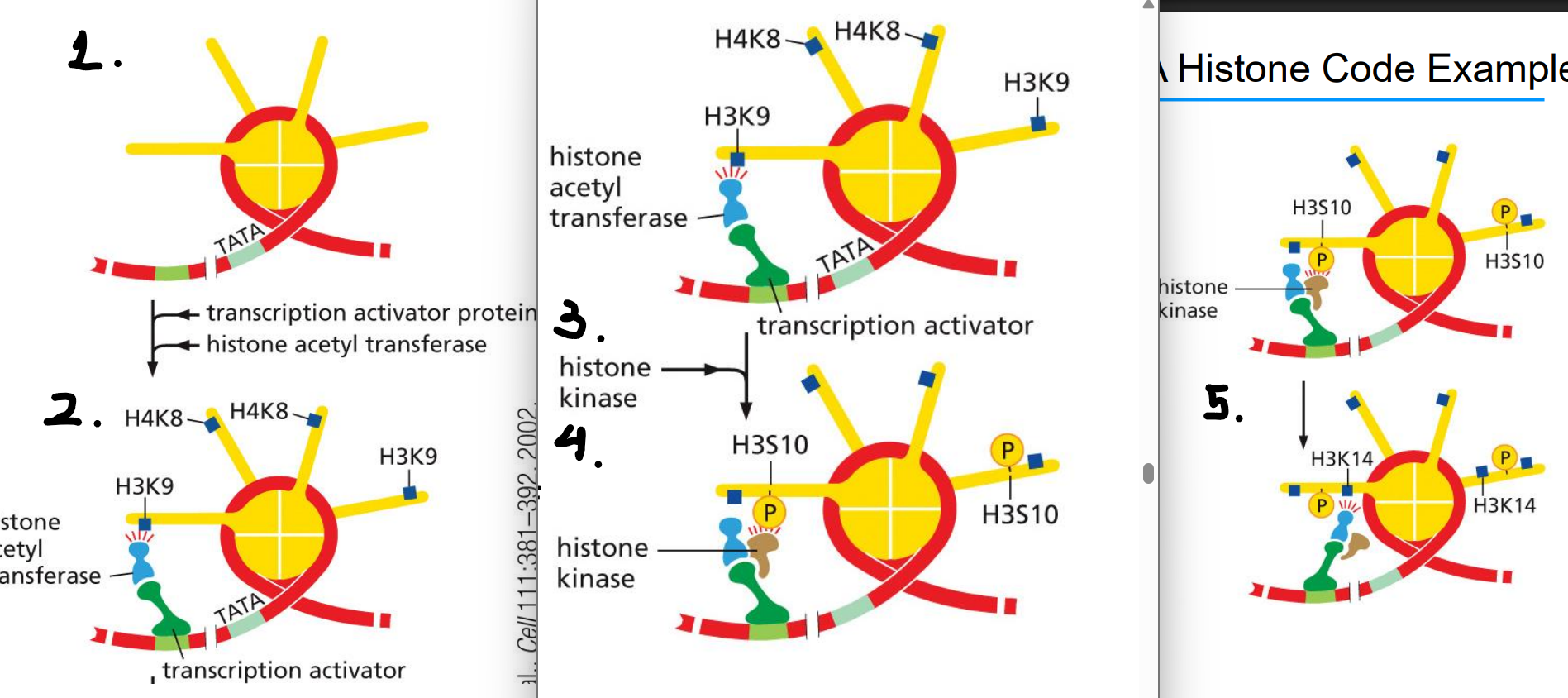
human interferon gene regulation as a histone code example
activator protein attracts histone acetyltransferase
HA acetylates H3K9 and H4K8
activator protein attracts histone kinase
histone kinase phosphorylates H3S10 (can only occur after acetylation of H3K9)
serine phosphorylation signals the acetyltransferase to acetylate H3K14
TFIID and a chromatin remodeling complex bind to modified tails and initiate transcription
the code for transcription initiation is written
transcriptional repression
competitive DNA binding
masking the activation surface
direct interaction with the general transcription factors
recruitment of chromatin remodeling complexes
recruitment of histone deacetylases
recruitment of histone methyl transferase
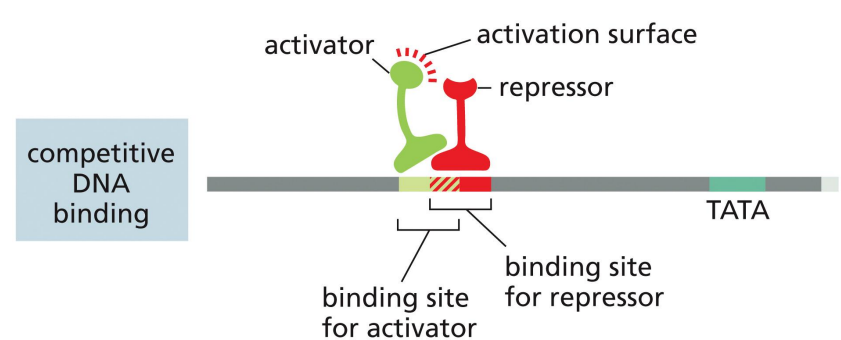
repressor proteins: competitive DNA binding
a mechanism of transcriptional repression where activator and repressor compete to bind to DNA (occurs if binding sites overlap)
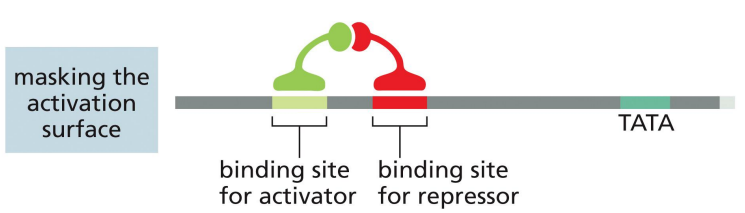
repressor proteins: masking the activation surface
a mechanism of transcriptional repression where the repressor prevents the activator’s activation surface from binding with transcription machinery
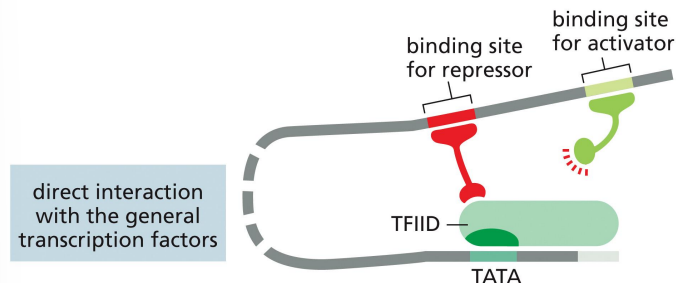
repressor proteins: direct interaction with the general transcription factors
a mechanism of transcriptional repression where the repressor interacts with the general transcription factors to prevent proper transcription
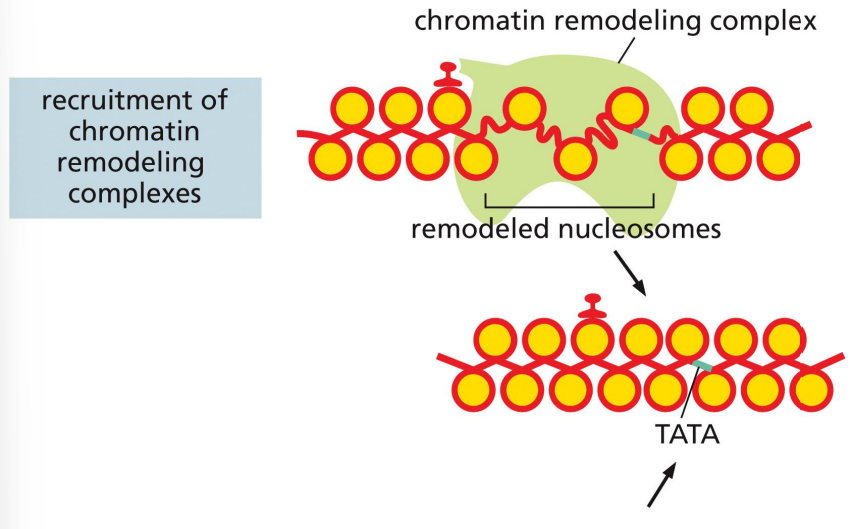
repressor proteins: recruitment of chromatin remodeling complexes
a mechanism of transcriptional repression where the repressor recruits the chromatin remodeling complex to restrict access to the promoter sequence
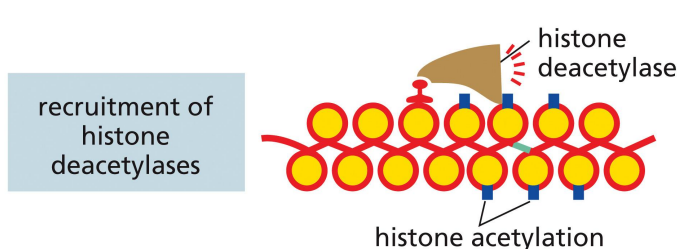
repressor proteins: recruitment of histone deacetylases
a mechanism of transcriptional repression where the repressor recruits histone deacetylases to interfere with histone code for transcriptional activation
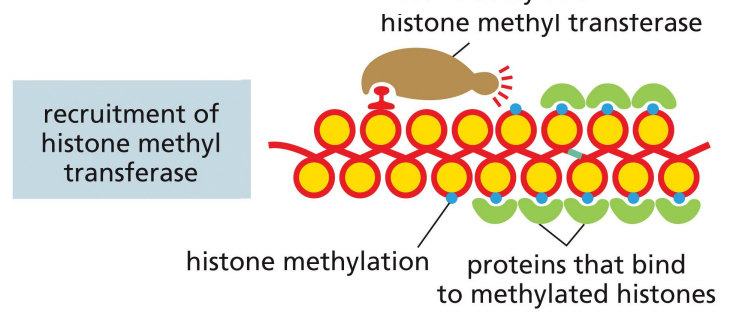
repressor proteins: recruitment of histone methyl transferase
a mechanism of transcriptional repression where the repressor recruits histone methyl transferase to interfere with histone code for transcriptional activation
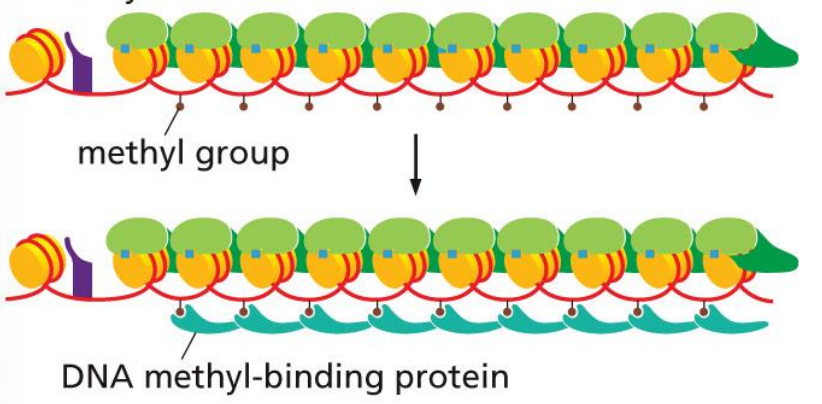
proteins can bind to these methylated histones that further compact chromatin
epigenetic inheritance
a process where gene expression patterns can be inherited by daughter cells, maintaining specialization across a lineage of cells
occurs by DNA methylase and DNA methyl-binding proteins that create inheritable structures
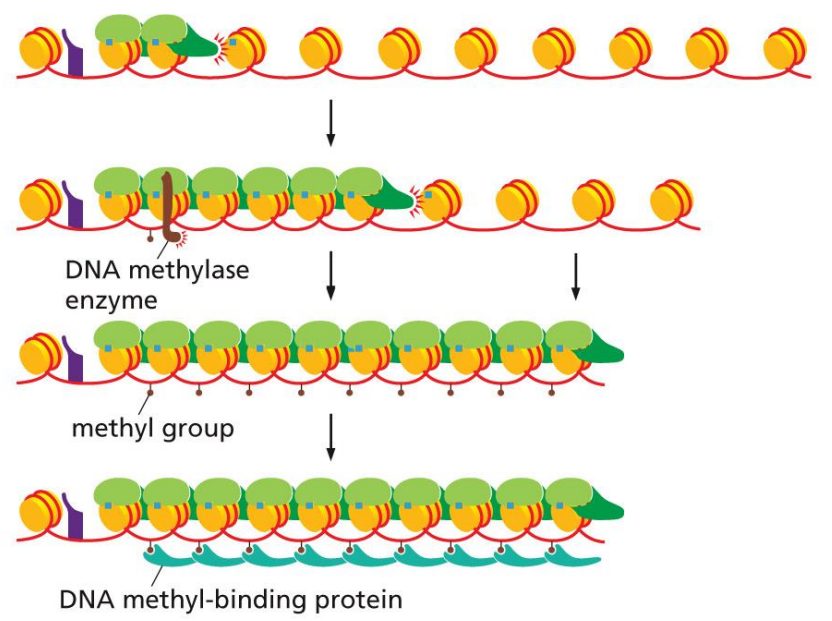
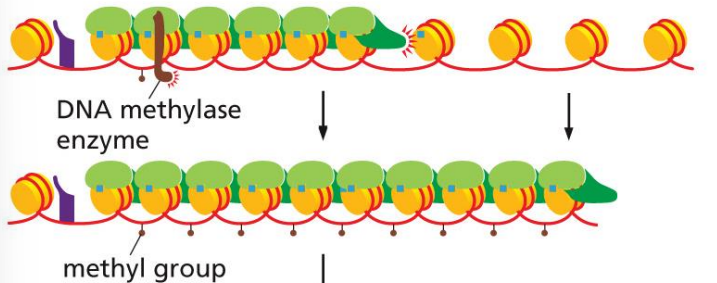
DNA methylase enzyme in epigenetic inheritance
attracted by reader and methylates nearby cytosines
the reader-writer chain reaction (one reader for every next histone) allows DNA methylase enzyme to methylate several linker DNAs in a row
methylation can be inherited in daughter cells
DNA methyl-binding proteins in epigenetic inheritance
bind methyl groups and stabilize structure, repressing genes that may not be necessary for certain specialized cells
because methylation is inheritable, these proteins will be attracted again to same genes in daughter cells
reader-writer complexes
writers are bound by transcription regulator and can recruit a reader protein
sets off a chain reaction where readers attract writers, and writers create a signal to attract readers
this complex spreads the histone code along chromatin and can stabilize it
DNA methylase and DNA methyl-binding proteins work together to further stabilize this structure, creating an inheritable gene expression pattern