Molecular genetics
1/51
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
52 Terms
nucleoside
sugar + base
nucleotide
sugar + base + phosphate(s)
purines
adenine and guanine
pyrimidines
cytosine + uracil + thymine
molecule of nucleotide and bonds within
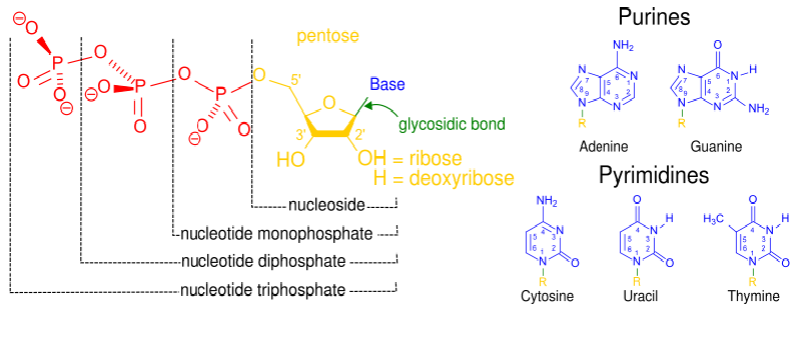
structure of DNA
Chain of deoxyribonucleotides - ACTG
Sugar phosphate backbone = sugars connect bases between 3' 5' carbons
Two polynucleotide chains run anti-parallel and pair up AT CG
Chains twist into double helix
double helix and its qualities to account for key functions of DNA
Replication = strict base pairing gives simple mechanism for making exact copy
Storage of info = order of bases form triplet genetic code (codon) carrying instructions to make specific protein
Stability = strong phosphodiester bonds keep sequence intact (weak H bonds allow message to be read when necessary)
gene
Genes are ordered sequence of nucleotides (DNA) that encodes a specific functional product - RNA or PROTEIN
3 types of RNA
rRNA and tRNA functional molecules in their own right
mRNA - immediate molecule carrying the instructions for making a specific protein
DNA --> mRNA --> Protein
where is genetic variation
At molecular level, visible variation is due to differences in our proteins which in turn is due to differences in our DNA sequence
To study genetic variation we can analyse proteins or DNA
what is genetic variation
Alterations to DNA sequences can stop or alter protein function --> disease / altered characteristics
Studying protein reveals underlying genetic variation (polymorphism)
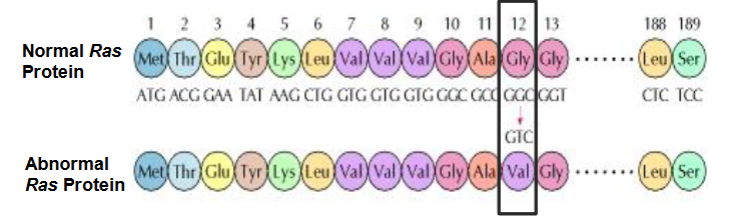
what changes don’t alter protein coded for
Redundant changes
Both codons code for same amino acid e.g. CGC and AGG for arginine
3rd base wobble
e.g. GGG GGA GGT GGC code for glycine
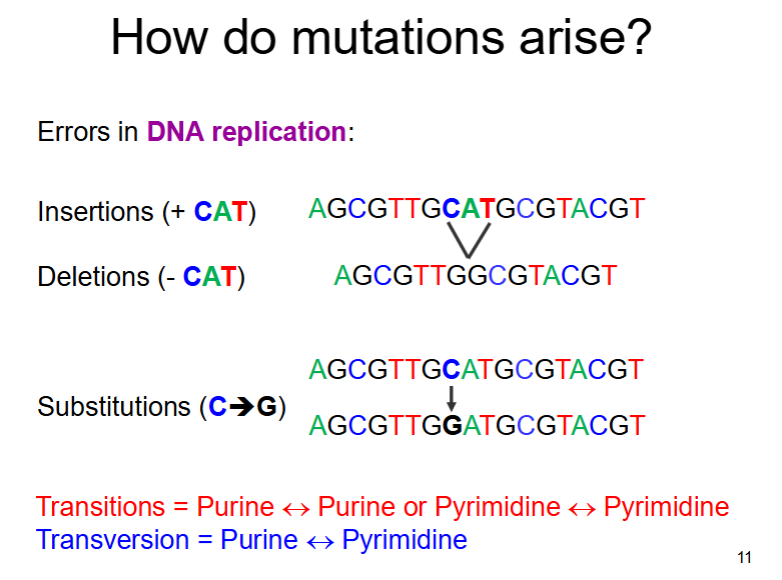
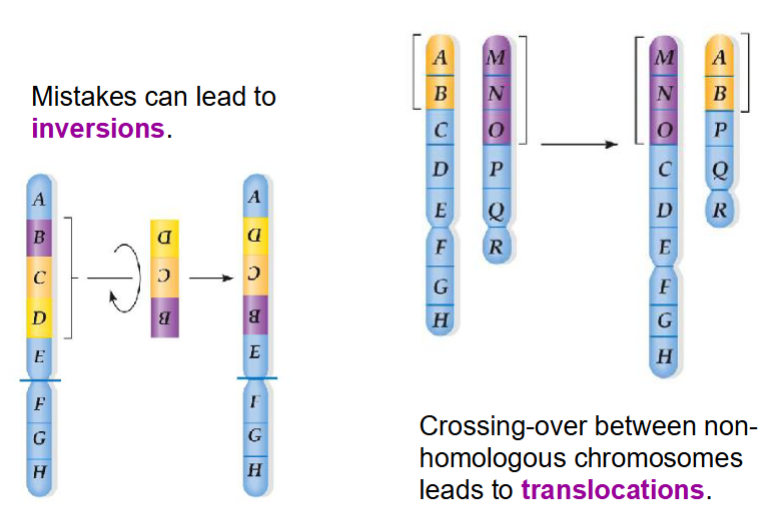
types of mutation - change in DNA sequence
- deletions
-duplications
-inversions
-insertions
-translocations
-substitutions
Can involve any number of nucleotides from single bases to large sections of chromosome
errors in DNA replication can cause mutations as follows:
errors in recombination (crossing over) during meiosis can cause mutations as follows:
Inversions, insertions or deletions between homologous chromosomes
Translocations between non-homologous chromosomes
meiosis and homologous chromosomes during crossing over
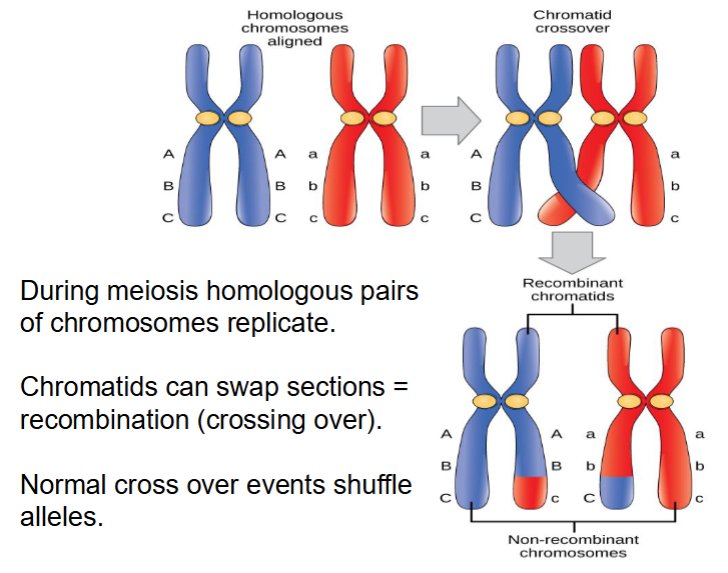
mistakes when crossing over can lead to which types of mutations?
insertions and deletions and duplications
visual of crossing over
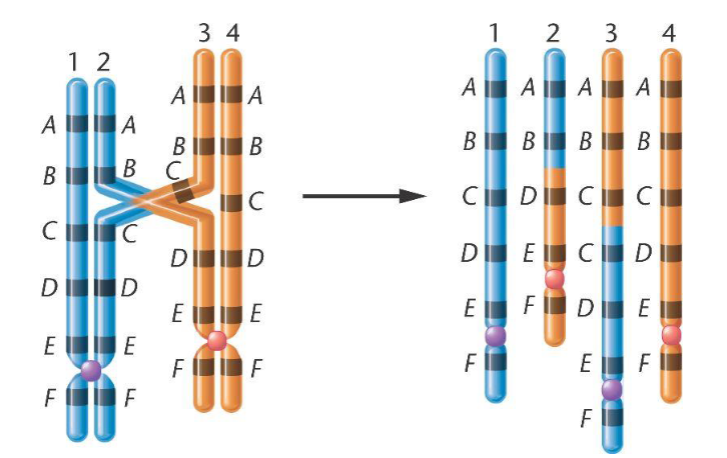
visual of crossing over leading to mutations
DNA analysis methods
If there are changes that do not alter a protein we need to analyse the genetic material (DNA) directly
DNA extraction
Identify the different variants (alleles)
Detect the results
where in a eukaryotic cell can DNA be extracted from?
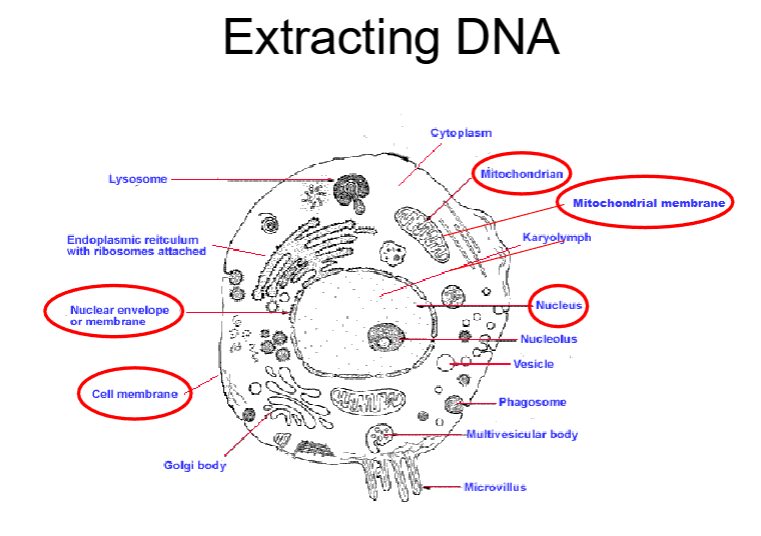
three main steps of DNA extraction
Cell lysis
A combination of heat, changes of pH, chemicals (e.g. detergents) and enzymes (proteases) are used to target proteins in cell membrane
Remove proteins
Enzymes, chemicals and separation by centrifugation can do this
Recover the DNA
Done by precipitation (DNA is insoluble in alcohol) or the use of specialised compounds that bind to the DNA
gel electrophoresis
DNA extracted and placed and placed in gel
Genomic DNA or isolated fragments can be analysed using electrophoresis
Gel matrix acts like sieve to separate fragments
examples of gels used in gel electrophoresis
Agarose (AGE)
Polyacrylamide (PAGE)
SDS-PAGE
electrophoresis
AGE or PAGE can be used to view DNA
DNA has overall -ve charge
Will move to anode
Small fragments = faster
Detect by staining with fluorescent dyes
distance travelled is …
equal to the length of DNA molecule (base pairs)
brightness of band is…
the amount of DNA present (ug)
locus
Specific point or location on chromosome (plural = loci)
allele
Different variations found at particular locus
polymorphism
Occurrence of more than one allele at a locus
genotype
Combination of alleles at locus
types of polymorphism - sequence variation
base substitutions e.g. SNPs - single nucleotide polymorphisms
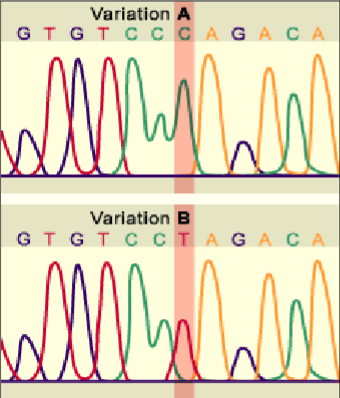
types of polymorphism - length variation
e.g. repetitive sequences
VNTRs - variable number tandem repeats
STRs - short tandem repeats
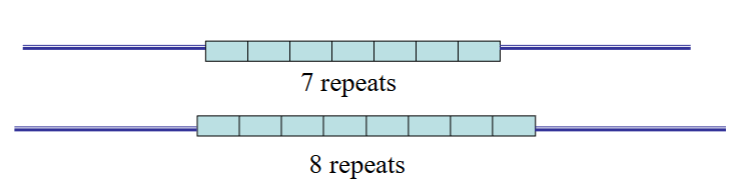
tools for molecular genetics - restriction enzymes
Type ii restriction endonucleases
-enzymes that cut within a molecule of nucleic acid
-each enzyme has specific recognition sequence
Polymorphisms analysed using restriction enzymes are known as RFLPs
Restriction Fragment Length Polymorphisms
each restriction enzyme has a specific recognition sequence for where they need to cut
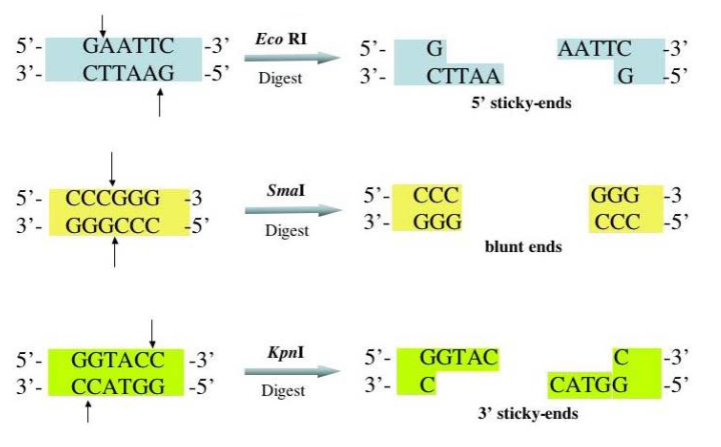
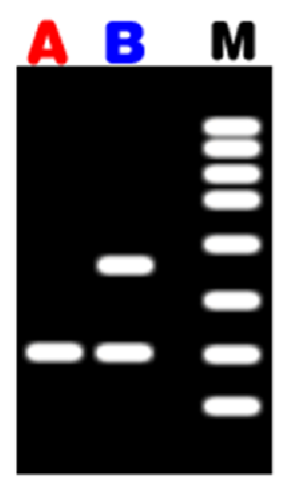
length variation can be detected e.g. analysis of VNTRs
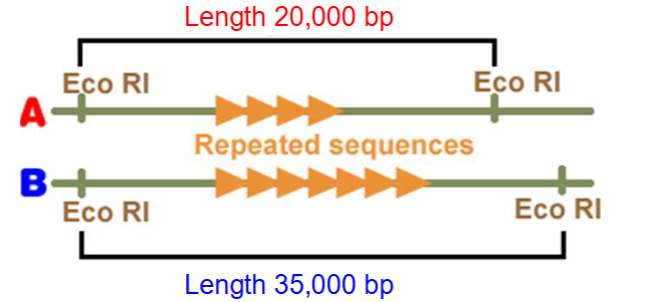
VNTRs
Enzyme cuts wither side of repeat region
Different DNA fragments are separated by electrophoresis
Length of fragments depends on number of repeats between two restriction sites
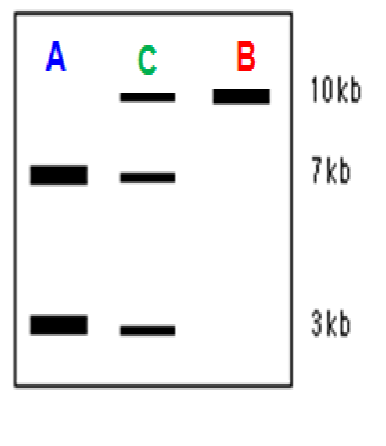
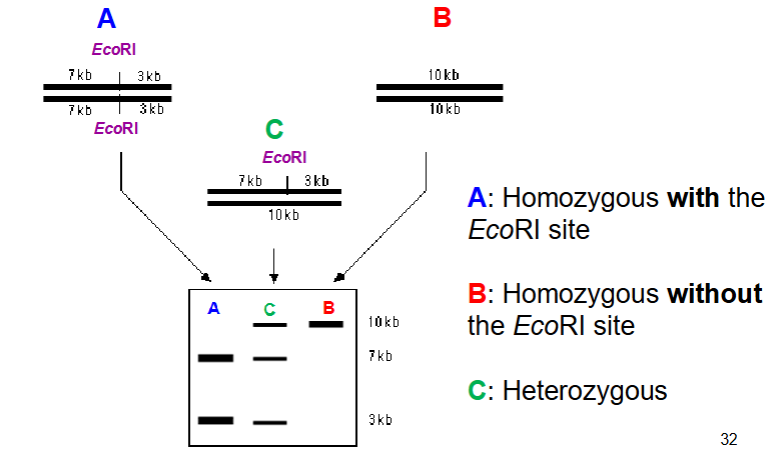
One band - homozygous
Two band - heterozygous
changes in DNA sequence (SNPs) can alter restriction enzyme recognition sites
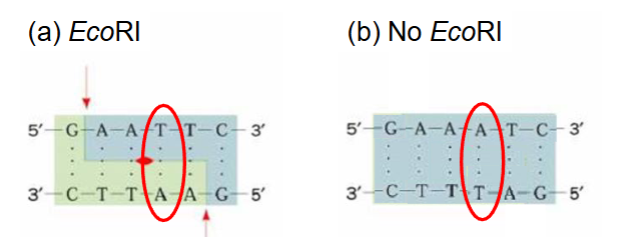
SNPs
DNA is digested with enzyme
Different DNA fragments are separated by electrophoresis
Number of bands depends on how often restriction enzyme cuts - revealing underlying genetic variation
looking at SNPs using restriction enzymes
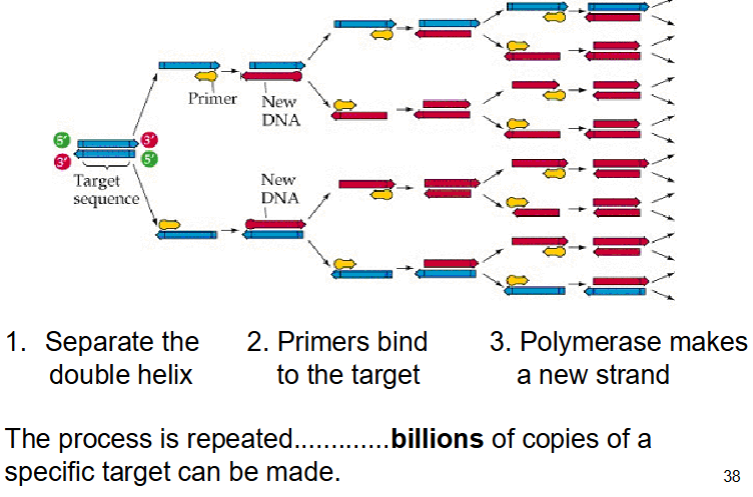
PCR - polymerase chain reaction - history
Kary Mullis first described PCR in 1985, he won Nobel prize for Chemistry in 1993
PCR allows us to isolate and copy a specific section of DNA
PCR reagents
Template DNA - to be amplified
dNTPs (deoxyribonucleotide triphosphates) - building blocks of DNA
Buffers - maintains optimum pH
DNA polymerase - the enzyme that synthesises DNA
DNA primers - define the region to be amplified - short single stranded bit of DNA , marks start and end point of sequence wanted to copy
Magnesium - cofactor required by enzyme
PCR process
forensic DNA analysis
All cells (except RBCs) contain nucleus so could provide DNA for analysis
PCR is very sensitive - only trace amounts of material needed
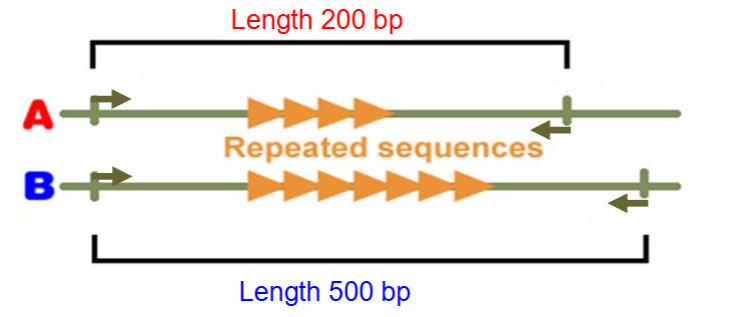
Forensic DNA analysis of STRs
Perfect for PCR based analysis
Similar structure to VNTR
But these are very short - 2-6 bases
DON’T use restriction enzymes - use PCR primers and PCR to copy

forensic DNA analysis of STRs
Copy repeat region (using PCR)
Length of each fragment depends on number of repeats - revealing the underlying genetic variation
Separate the fragments using electrophoresis and measure the lengths
Identify the alleles a person carries and their genotype
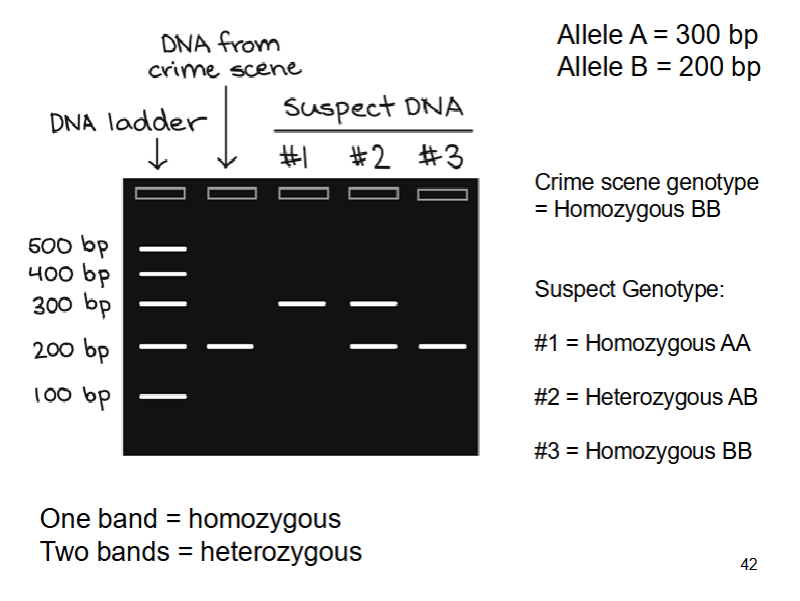
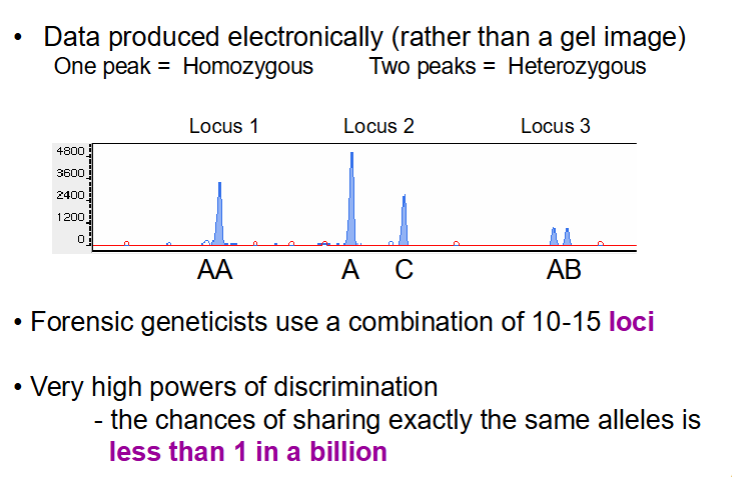
forensic DNA analysis of STRs
Don’t tend to use agarose gels but rather electronic machinery
Produce peaks
1 peak = 1 band on gel
forensic genetics - from crime scene to court
Identification of particular genetic variants (alleles) present in a suspect and scene of crime evidence
Is there a match?
Apply population genetic theory to give courts a measure of probability
Missing person and DVI
Identify matching body parts
Identify individual by matching DNA to personal items
Identify individual by comparing DNA to a known relative
paternity testing
Mendelian genetics determines that a child must share one allele with each parent
Genetic testing can include or exclude a tested man from being the biological father of a child
mendelian genetics
Molecular genetic analysis allows us to reveal variation in our DNA
Mendelian genetics determines patterns of inheritance - a child must share one allele with each parent
Combo of alleles you inherit (genotype) determine characteristics expressed (phenotype)
population genetics
Q: how likely is it that the DNA match/paternity could be attributed to someone other than suspect?
When we present DNA evidence in court we need to understand probabilities
This requires estimates of allele/genotype frequency and the application of population genetic theory