GB1- CHapter 17
1/34
Earn XP
Description and Tags
Gene Expression
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
35 Terms
Archibald Garrod
proposed that genes control enzymes to do specific chemical reactions, ultimately affecting phenotype
thought that inherited diseases came from an inability to synthesize a certain enzyme in the metabolic pathway
ex) Alkaptonuria- black urine due to inability to break down tyrosine and phenylalanine
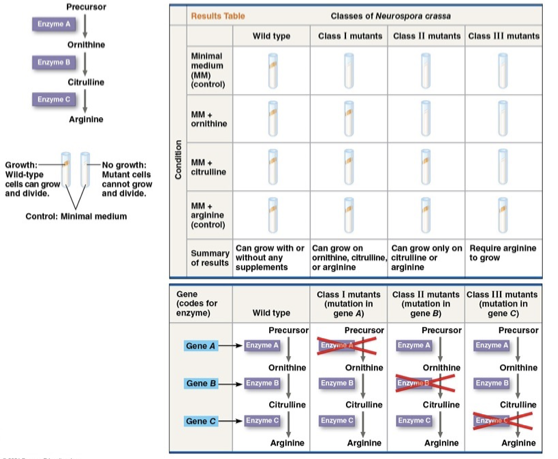
George Beadle and Edward Tatum
made Neurospora Crassa (bread mold) mutants by exposing them to X Ray so that it can’t survive off just the minimal medium. The mutants were placed in minimum mediums+ one nutrient, and whichever nutrient presence caused it to grow, was missing from the mutant.
Adrian Srb and Norman Horowitz
identified three arginine deficient mutants that each lacked a different enzyme to synthesize arginine
“One gene One enzyme hypothesis”
One gene one enzyme hypothesis
hypothesis that a particular gene directs a particular enzyme
not all proteins are enzymes, so it was renamed to “One gene, one protein hypothesis”. proteins are made up of multiple polypeptides, so it was then renamed to “One gene, one polypeptide” hypothesis.
Transcription
the synthesis of mRNA using information in DNA
Translation
polypeptide synthesis using mRNA information
site of translation: ribosomes
central dogma
flow of information from DNA to RNA to protein
# of amino acids, # of nucleotide bases
20 amino acids, 4 nucleotide bases
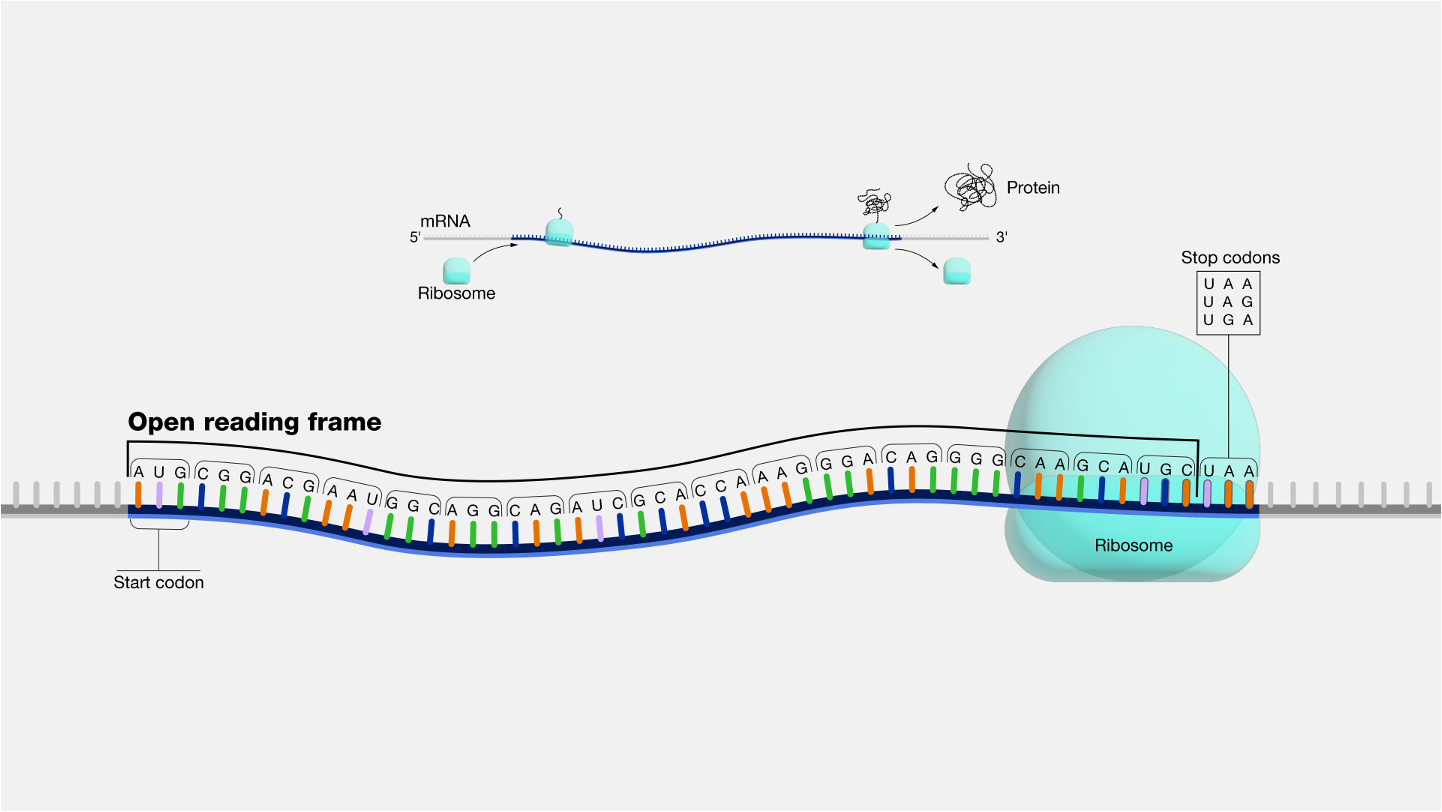
codon
nonoverlapping, three nucleotide letters on mRNA that translate to amino acids
start codon- AUG
64 codons- 61 for amino acid coding
3 stop codons UAA UAG UGA
redundant but not ambiguous. Multiple codons can code one amino acid, but each codon only codes for one amino acid.
reading frames- correct groupings of codons
open reading frame- regular transcription mRNA with start and stop codons
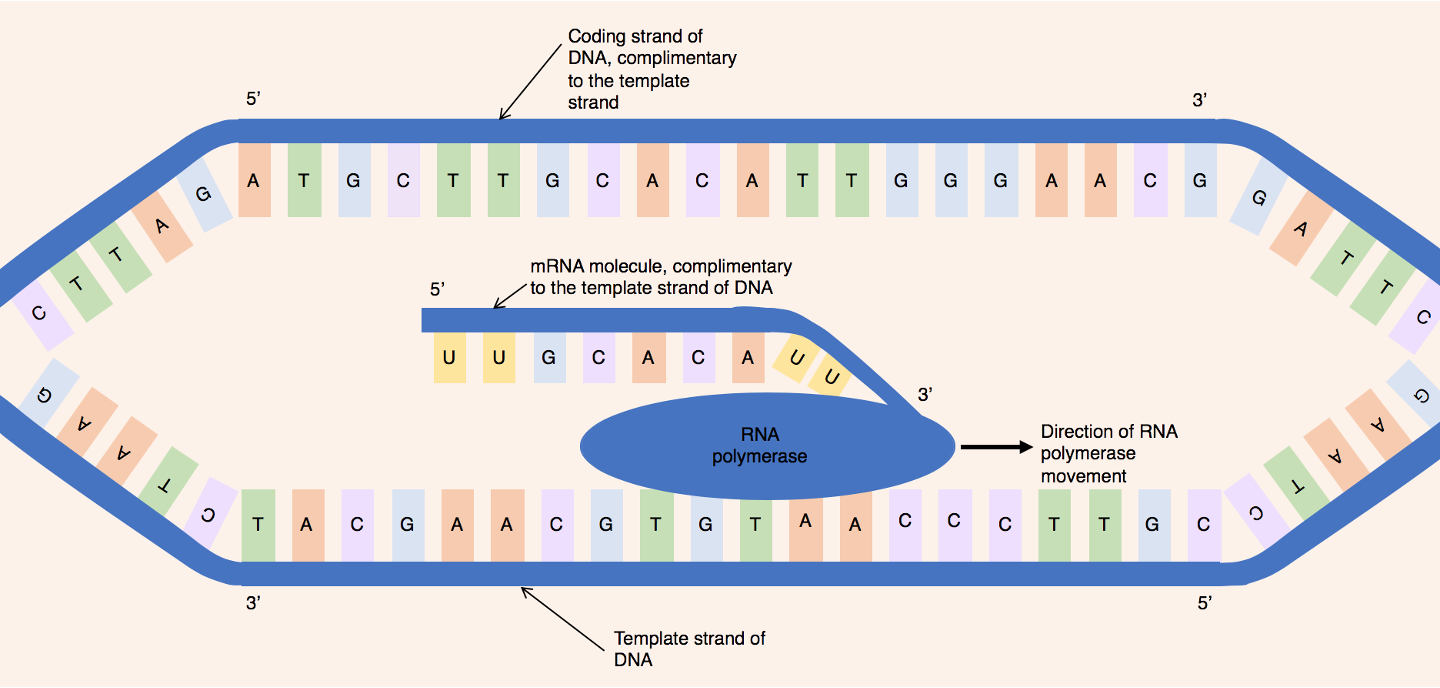
template strand
strand used as a template in transcription
template strand is determined by a specific nucleotide sequence
template strand is always the same strand for a particular gene
opposite strand further away may be used as a template strand for another gene
coding strand
non template strand that has identical code as codons (T in place of U)
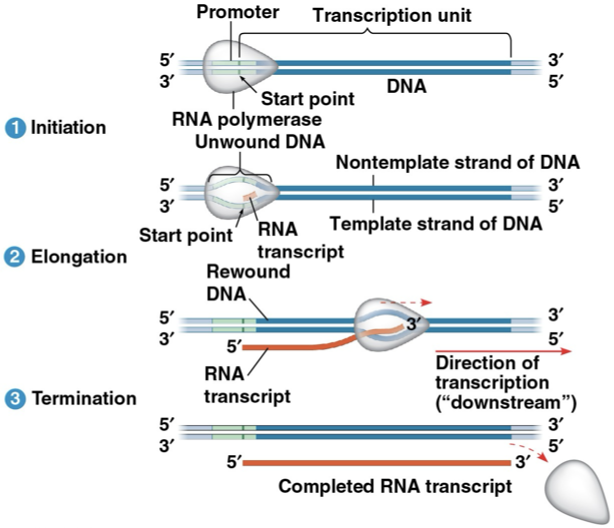
Bacteria Transcription
transcription ends at “terminator”
transcription unit- the stretch of DNA being transcribed
RNA synthesis
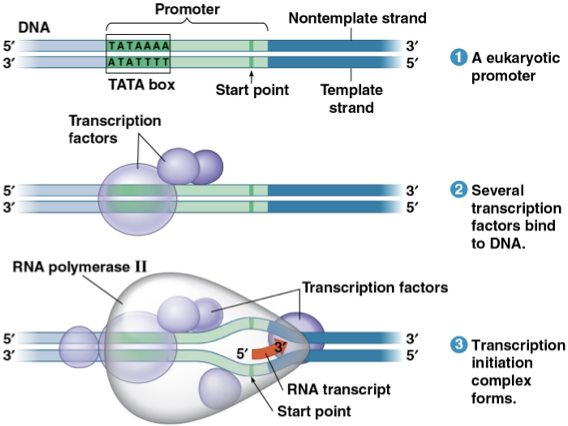
Initiation- RNA polymerase II latches onto the promoter with the help of transcription factors. In eukaryotes, there’s a TATA box. The RNA polymerase II pries the two strands of DNA apart and RNA replication begins at the “start point” of the template strand
Elongation- The DNA strand being copied elongates
Termination- RNA polymerase II falls off after transcribing the polyadenylation signal, leaving behind a transcription unit
a gene can be transcribed simultaneously by several RNA polymerases
RNA polymerase doesn’t need a primer!
transcription initiation complex
compelete assembly of promoter, transcription factors, and RNA polymerase II
in eukaryotes, there is a promoter called TATA box
RNA processing
alteration of mRNA before it is sent to the ribose
5’ end gets a 5’ cap
3’ end gets a poly A tail- polyadenylation: synthesized by poly A polymerase
functions
facilitate the export of mRNA to the cytoplasm
protects mRNA ftom hydrolytic enzymes
helps ribosomes attach to the 5’ end
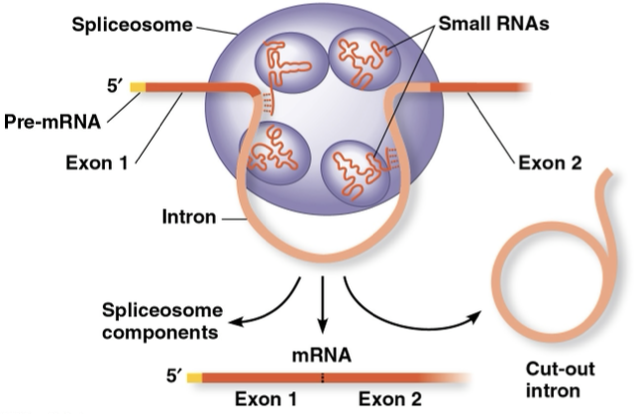
RNA splicing
removal of introns, and noncoding segments, by spliceosomes
spliceosomes- proteins and several small RNA that recognize splice sites
exons- translated into amino acids and expressed
Ribozymes
catalytic RNA molecules that can splice RNA
properties that allow it to function as an enzyme
Able to form 3D structure because it can base pair by itself
Some RNA bases have functional groups that can catalyze
RNA can hydrogen bond with other nucleic acids
alternate RNA splicing
gene can give rise to multiple different polypeptides depending on which segments are considered as exons
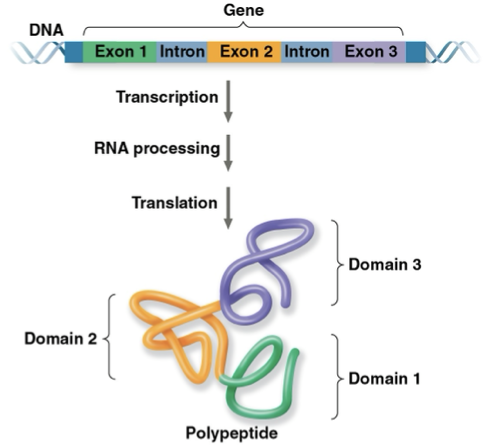
domains
different regions of proteins
different exons code for different domains
exon shuffling
mixing and matching of different exons of different genes that may result in new proteins
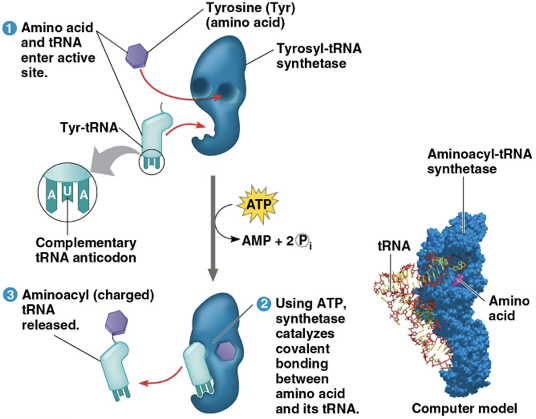
accurate translation requires two instances of molecular recognition
first: aminoacyl-tRNA synthetase correctly matches tRNA with amino acid
second: correct match between tRNA anticodon and mRNA codon
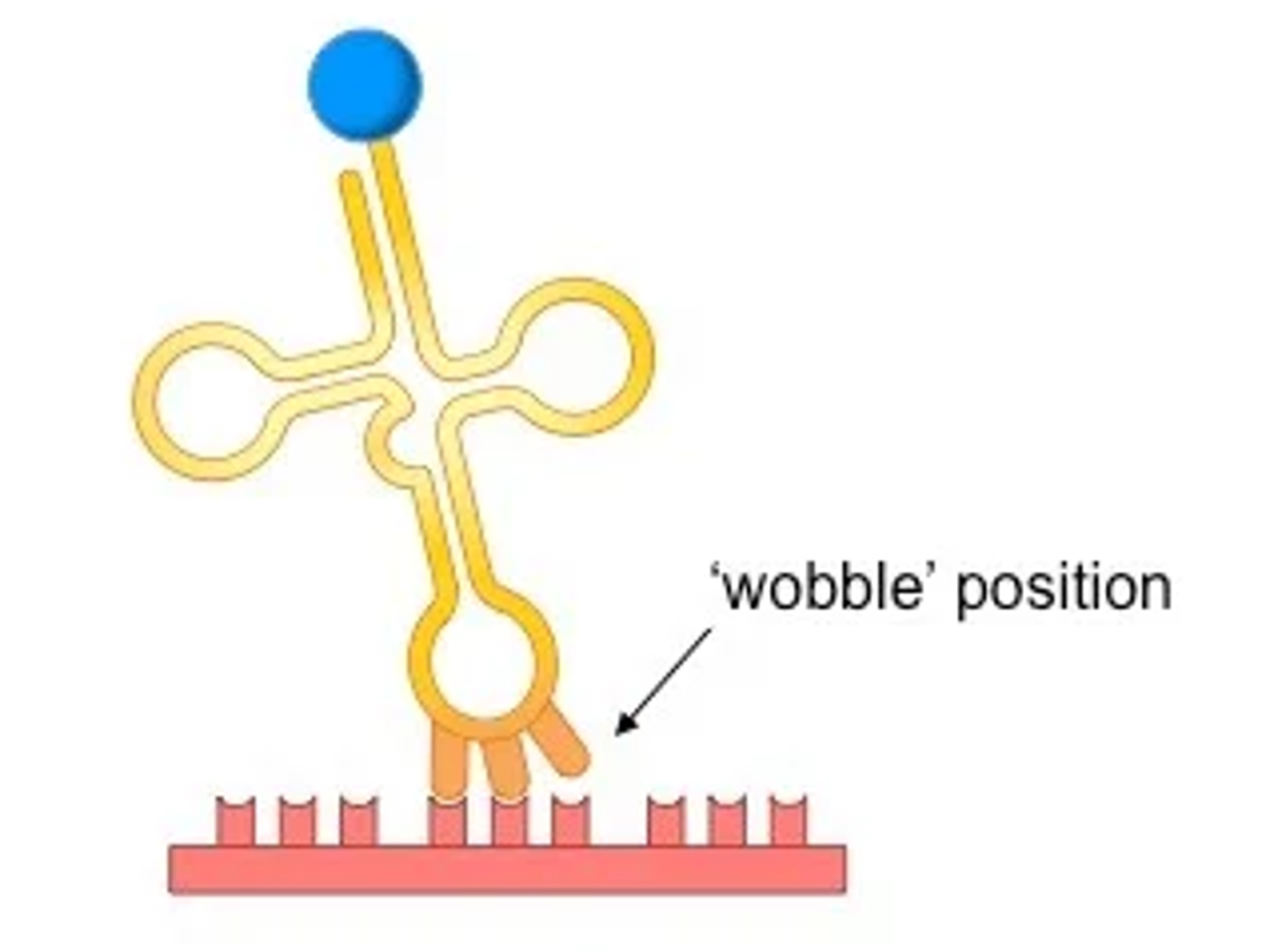
wobble
flexible base pairing of the third base allows some tRNAs to bind to more than one codon
Ribosomes
made up of proteins and ribosomal RNAs
eukaryotic ribosome is bigger than prokaryotic and has a different molecular composition- allowing antibiotics to inactivate bacterial ribosomes without hurting the eukaryotic one
ribosomes facilitate the coupling of tRNA with mRNA codons
E site: where the tRNA leaves
P site: holds the polypeptide chain
A site: incoming tRNA and amino acid docking site
translation steps
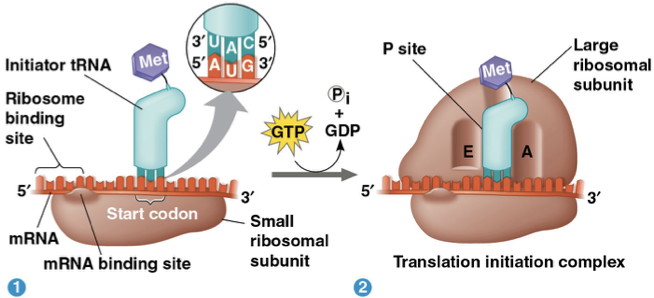
initiation- small ribosomal subunit binds with mRNA and indicator tRNA that carries amino acid methionine. The small subunit moves along the mRNA until it reaches the start codon AUG. Initiation factors bring in the large subunit that completes the translation initiation complex.
elongation- amino acids are added one by one to the C terminus of the growing chain with the help of elongation factors
codon recognition- GTP required: anticodons on tRNA correctly bind to mRNA codons
peptide bond formation- rRNA of large ribosomal subunit forms polypeptide bonds between carboxyl ends of amino acids. The polypeptide chain is transferred from P site to A site tRNA.
translocation- GTP required: A site tRNA moves over to P site and P site tRNA exits through the E site.
Termination- stop codon is reached and release factor binds to site A and binds water instead of amino acid. The hydrolysis reaction causes the whole ribosomal subunit to fall apart.
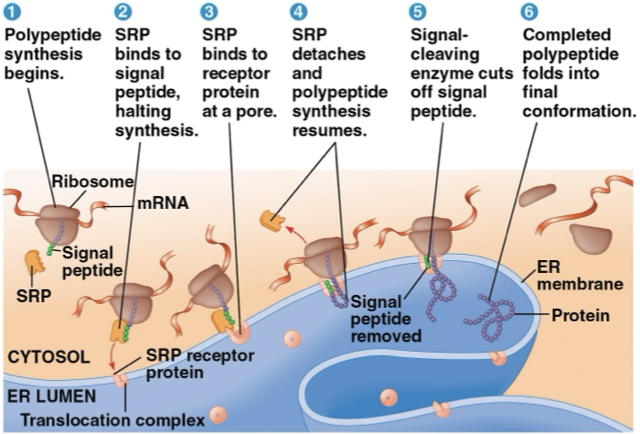
targeting functional protein
free ribosome- in the cytosol and synthesizes proteins that function in the cytosol
bound ribosome- bound to the ER and synthesizes proteins that function in the endomembrane system or secreted
synthesis starts and finishes in the cytosol unless polypeptide signals attach
signal peptide- attaches to polypeptide and signals it to go to the ER
signal recognition particle (SRP)- binds to signal peptide and escorts it to the ER
the signal peptide is removed by the enzyme
polypeptides are directed to other organelles
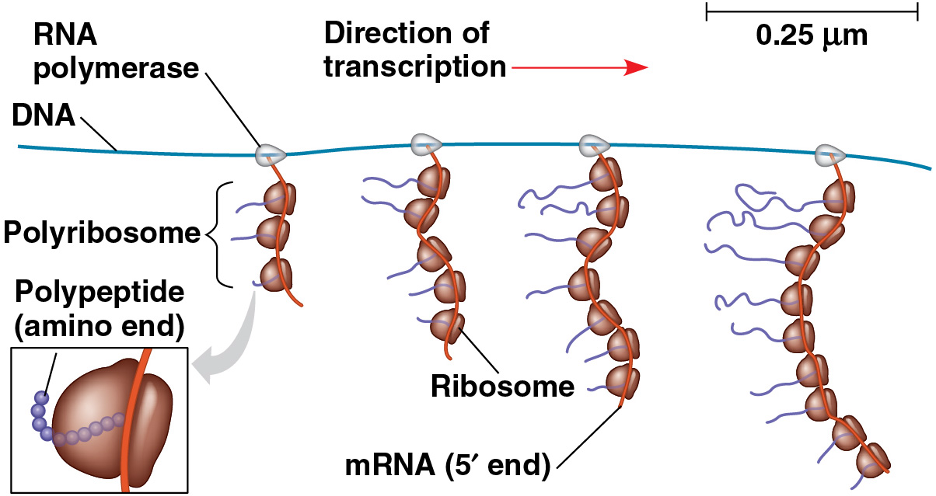
polyribosome/ polysome
enables a cell to make many polypeptide copies very quickly
in prokaryotes, the whole process is even faster because transcription and translation can occur almost simultaneously
mutations
changes in the genetic information of the cell
point mutations- changes in one nucleottide PAIR of a gene
single nucleotide pair substitutions
nucleotide pair insertions or deletions
silent mutations
no effect on amino acid produced by a codon because of redundancy in the genetic code
missense mutations
still code for an amino acid, but not the correct amino acid
Nonsense mutations
change an amino acid codon into a stop codon; most lead to a nonfunctional protein
frameshift mutation
Insertion or deletion of nucleotides (not pairs) may alter the reading frame
mutagens
physical or chemical agents that can cause mutations
Most carcinogens (cancer-causing chemicals) are mutagens, and most mutagens are carcinogenic
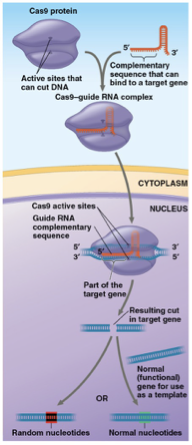
gene editing
altering genes in a specific way
CRISPR-Cas9 protein Cas9 is guided by guide RNA that leads to a target gene. Cas9 will cut both ends of the segment that’ll trigger a DNA repair system where the repair enzymes will add or remove random nucleotides- disabling a given gene
A template can be introduced with a normal copy of the gene that can be corrected