21: Microevolution-Genetic Changes w/in populations
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
microevolution
- describes small-scale genetic changes in population
- responds to shifting environmental circumstances
- ex : antibiotic resistance in bacteria
- study via analyzing variations within natural population and how and why these variations are inherited
macroevolution
- describes larger-scale evolutionary changes in a species and more inclusive groups
- occurs at or above the level of the species
- can result in speciation or the emergence of new species
- results from gradual accumulation of microevolutionary changes
population
all individuals of a single species that live together in the same place and time
phenotypic variations
- all difference in appearance or function that are passed from gen to gen
- caused by : genetic differences, differences in environmental factors, and interactions bt genetics and environment
only _________ based variation is subject to evolutionary change
genetic
quantitive traits
- a measurable phenotype that depends on the cumulative actions of many genes and the environment
- able to be described on a bar graph or curve
qualitative traits
- trait that fits into discrete categories
- polymorphism : exiting in >= 2 discrete states
- described by percentage/frequency of each trait
- intermediate forms are often absent
- frequently controlled by one or just a few genes -> simply-inherited traits
processes that generate genetic variations
1) production of new alleles -> small-scale mutations
2) rearrangement of existing alleles into new combos -> larger scale changes in chromosome structure/# and from crossing over, independent assortment, and random fertilizations
how biologist detect genetic variations?
1) gel electrophoresis : inferring the genetic variation in gene coding for a protein -> separates >= 2 forms of a protein that differs significantly in shape, mass, or net electrical charge
2) DNA sequencing tech : allows direct survey of genetic variation; can help accumulate astounding knowledge of genome -> study of DNA polymorphisms
population genetics
- understanding the causes of genetic variation within populations
- describe genetic structure of population
- create hypotheses formalized in mathematical models to describe evolutionary proceses
- test the predications of these models to evaluate the idea about evolution
gene pool
- sum of gene copies (alleles) at all gene loci in all individuals in the population
allele
1 of 2 or more versions of DNA sequence at a given genomic location (locus)
genotype frequencies
percentages of individuals in a population possessing each genotype
- each diploid organism has two copies of each gene
- 2 alleles may be same or different
- 3 types, pp, pq, and qq (p is dom, q res)
- p^2 + 2pq + q^2 = 1
allele frequencies
- relative abundances of the different alleles
- can be calculated
- two types (p and q)
- if genotype has a unique phenotype -> incomplete dominance -> allele frequencies can be counted directly
- p + q = 1
null models
- theoretical reference points against which observations can be evaluated
- predict what would happen if a particular factors had no effect
hardy-weinberg principle
- mathematical model that described genotype frequencies for sexually reproducing organisms
- the population's allele and genotype frequencies remain constant unless there is some type of evolutionary force acting upon them
- genetic equilibrium only if all are met : (1) no mutations are occurring; (2) population is closed to migration; (3) population is infinite in size; (4) all genotypes survive and reproduce equally well; (5) individual mate randomly wrt genotypes
- if these conditions are not met -> change of allele frequencies -> microevolution
factors of microevolution
mutation, gene flow, genetic drift, natural selective, nonrandom mating
what is the major source of heritable variations
mutations
- note : little to no immediate effect but creates new genetic variations over time
- most animals -> only mutations in germ line are heritable
deleterious mutations
- alter an individual's structure, function, or behavior in HARMFUL ways
- ex : ehlers-danlos syndrome -> mutation in coding for collagen -> loose skin, weak joints
- dermatosparaxis ehlers-danlos syndrome -> connective tissue disorder -> fragile skin, bruising and scarring, hernias, puffy eyelids, etc
lethal mutations
- may kill all carriers (if dominant) or only homozygous carriers (if recessive)
- any mutation that causes an individual to die before it reaches its reproductive age
- ex huntington's disease (dominant); cystic fibrosis (res); sickle cell anemia (res)
neutral mutations
- neither harmful or helpful
- mostly arise due to change in 3rd position of a codon
- degeneracy of the genetic code -> still produce the same amino acid
- can be beneficial later if environment changes
advantageous mutations
- has benefit to the individual that carries it
- natural selection may preserve the new allele and increase its frequency over time
gene flow
- movement of organisms or their gametes (eg pollen), or genetic variation from one pop. to another (migration)
- immigrants reproduce -> introduce new alleles -> shift alleles and genotype frequencies away from hardy-weinberg model
- there must be reproduce of immigrants into the gene pool, not just the act of migrating
- evolutionary importance depends on : (1) degree of genetic differentiation bt populations -> can be beneficial or harmful; (2) rate of gene flow bt the populations
genetic drift
- chance events that cause allele frequencies in a population to change unpredictable
- founder effect : extreme ex; small group splits from main pop. and find new colony -> ex amish in PA and ellis-van creveld syndrome; alleles may be missing or rare alleles occur more highly
- population bottleneck : population is sharply reduced in size by natural disaster or animal poaching; ex elephant seals have no variation in 24 proteins; lack of more rare/low frequency alleles
- more common in small populations
- generally leads to loss of alleles and reduce genetic variability
relative fitness
- number of surviving offspring that an individual produces compared w/others in the population
- differences is the main source for natural selection -> natural selection has little to do w/genes affecting an individuals post-reproductive life
directional selection
- extremely common, used in artificial selection
- individuals near one end of the phenotype spectrum have the highest relative fitness
- shift the mean phenotype towards the end the distribution favored
- body size in guppies
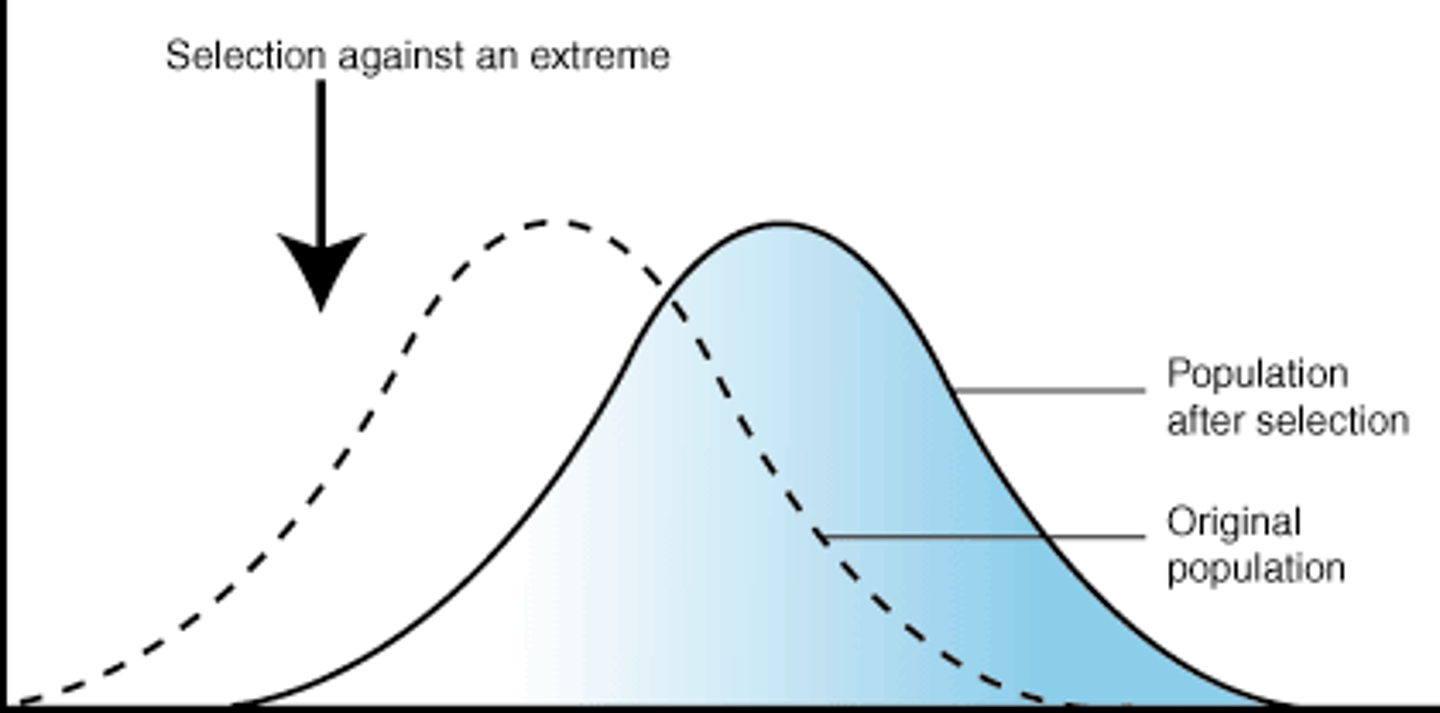
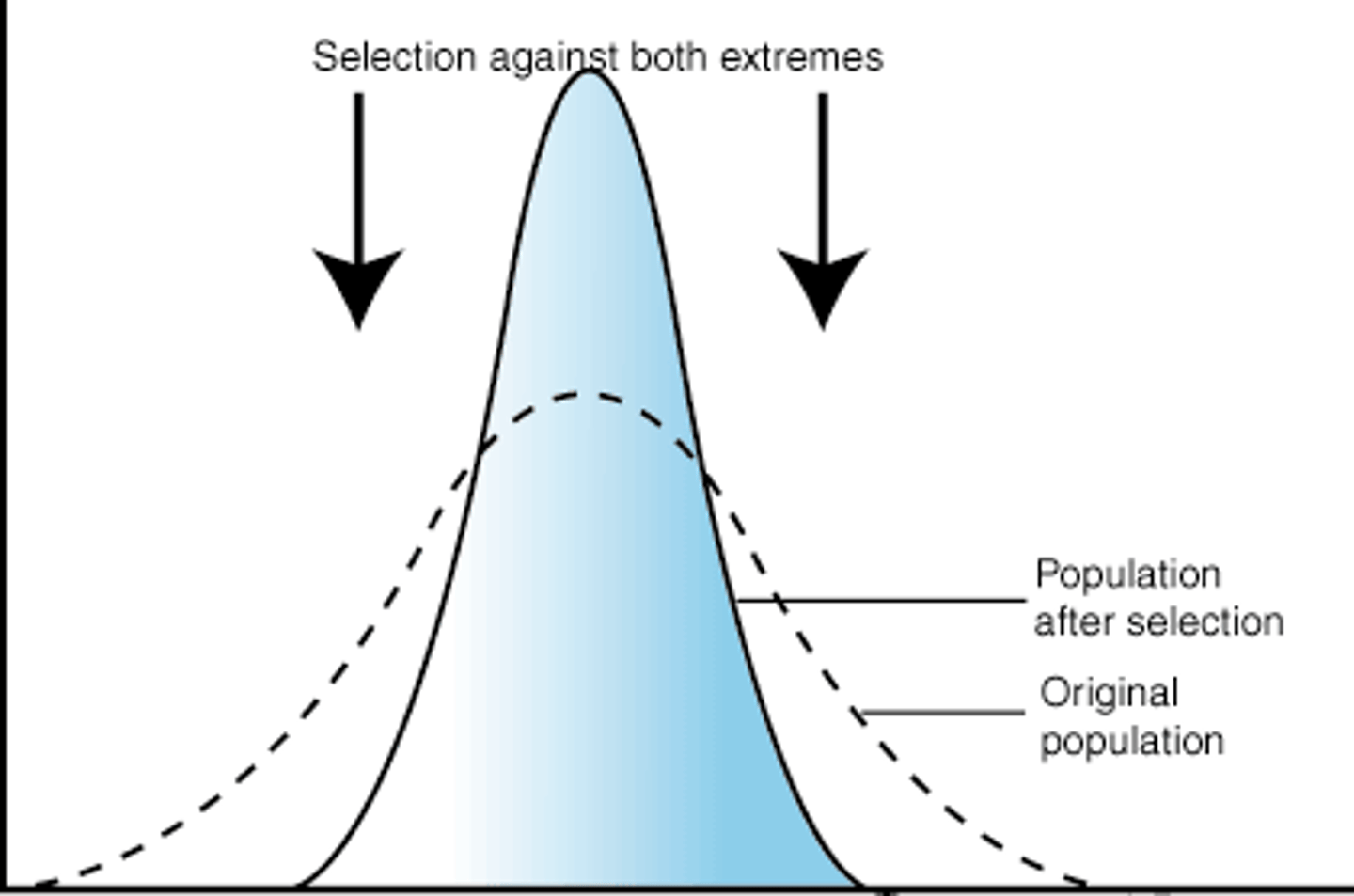
stabilizing selection
- when individuals expressing intermediate phenotypes have the highest relative fitness
- eliminated the phenotype extremes -> reduces phenotypic variation
- ex birthweights in human -> mean of 7-8lbs
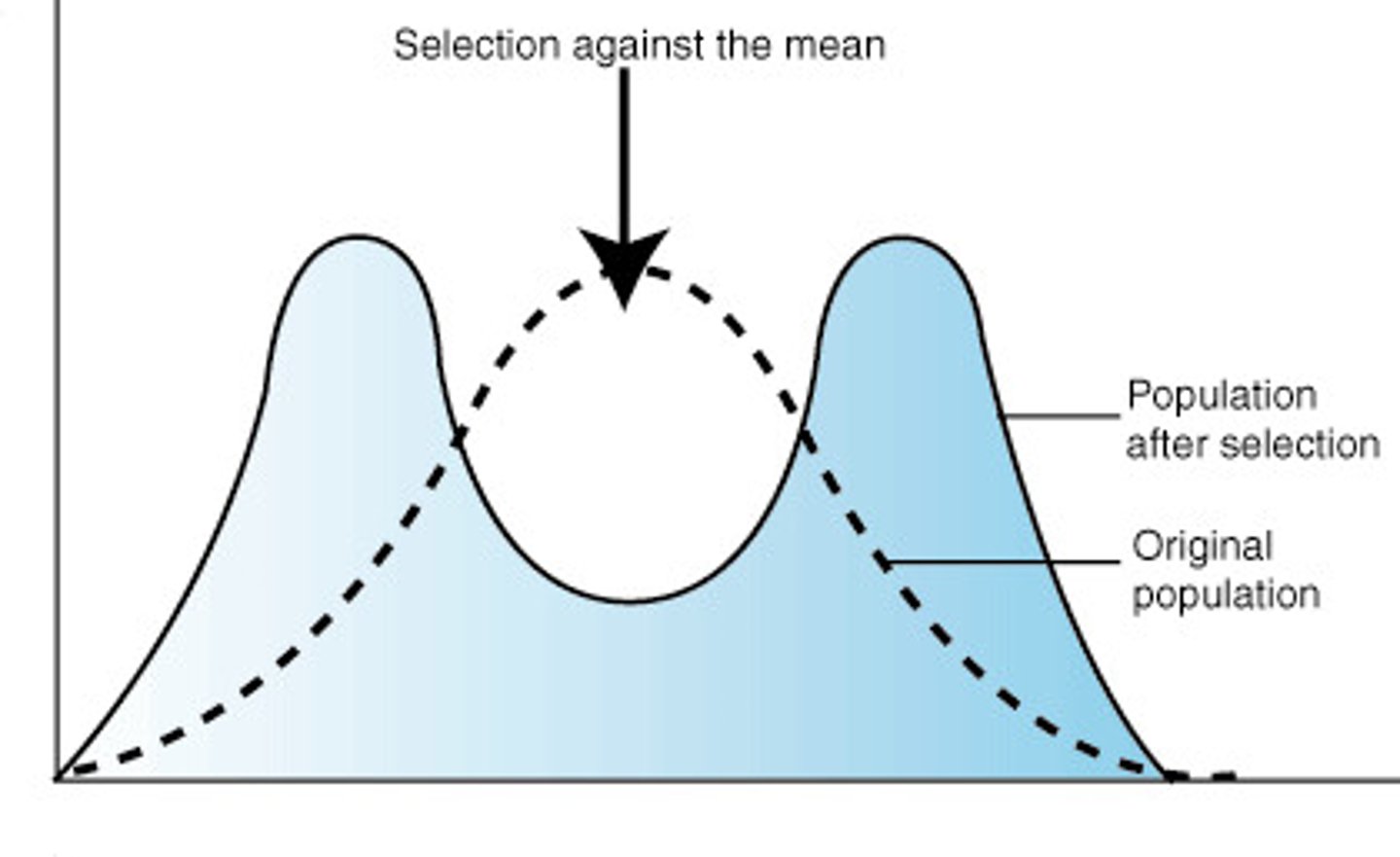
disruptive selection
- when extreme phenotypes have higher relative fitness than intermediate phenotypes
- promotes polymorphism
- least common
- ex bill size in medium ground finches
sexual selection
- special process that fosters evolution towards showy structures
- "special case" of natural selection
- acts on an individual's ability to obtain or successfully copulate w/a mate -> bright colors, elaborate courtship behaviors
- probable cause of sexual dimorphism (differences bt sexes in the same species)
intersexual selection
- based on interactions bt male and females
- males produce useless structures to make females attracted
- ex african male widowbird -> longer tail length
intrasexual selection
- based on interactions bt male and male
- males produce structures to intimidate, injure, or kill rival males
inbreeding
- form of nonrandom mating
- genetically related individuals mate w/each other
- self-fertilization is an extreme example
- recessive phenotypes are often expressed
balanced polymorphisms
- when 2 or more phenotypes or maintained in fairly stable proportions over gens.
- when heterozygotes have higher relative fitness, different alleles favored in different environments, or when rarity of phenotype provides an advantage (sickle cell and malaria)
frequency dependent selection
- genetic variability is maintained in a pop. bc a rare phenotype has a higher relative fitness -> becomes the more common phenotype -> rare phenotype loses its advantage
- ex bees pollinating elderflowers w/ color polymorphism -> rarer the color, the higher the reproductive success or higher relative fitness
neutral variations
- some genetic variation at loci coding for enzymes and other soluble proteins is selectively neutral (when different forms of the proteins function equally well)
- directly proportional to population size and length of time over which variations have accumulated
adaptive trait
- any product of natural selection that increases the relative fitness of an organism in its environment
- crucial for species survival and evolution of new species
- not all traits are adaptive
- adaptive changes in a organism's morphology must be based on SMALL modifications of existing structures
adaptation
- accumulation of adaptive traits over time