Protein Synthesis in Prokaryotes
1/48
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
49 Terms
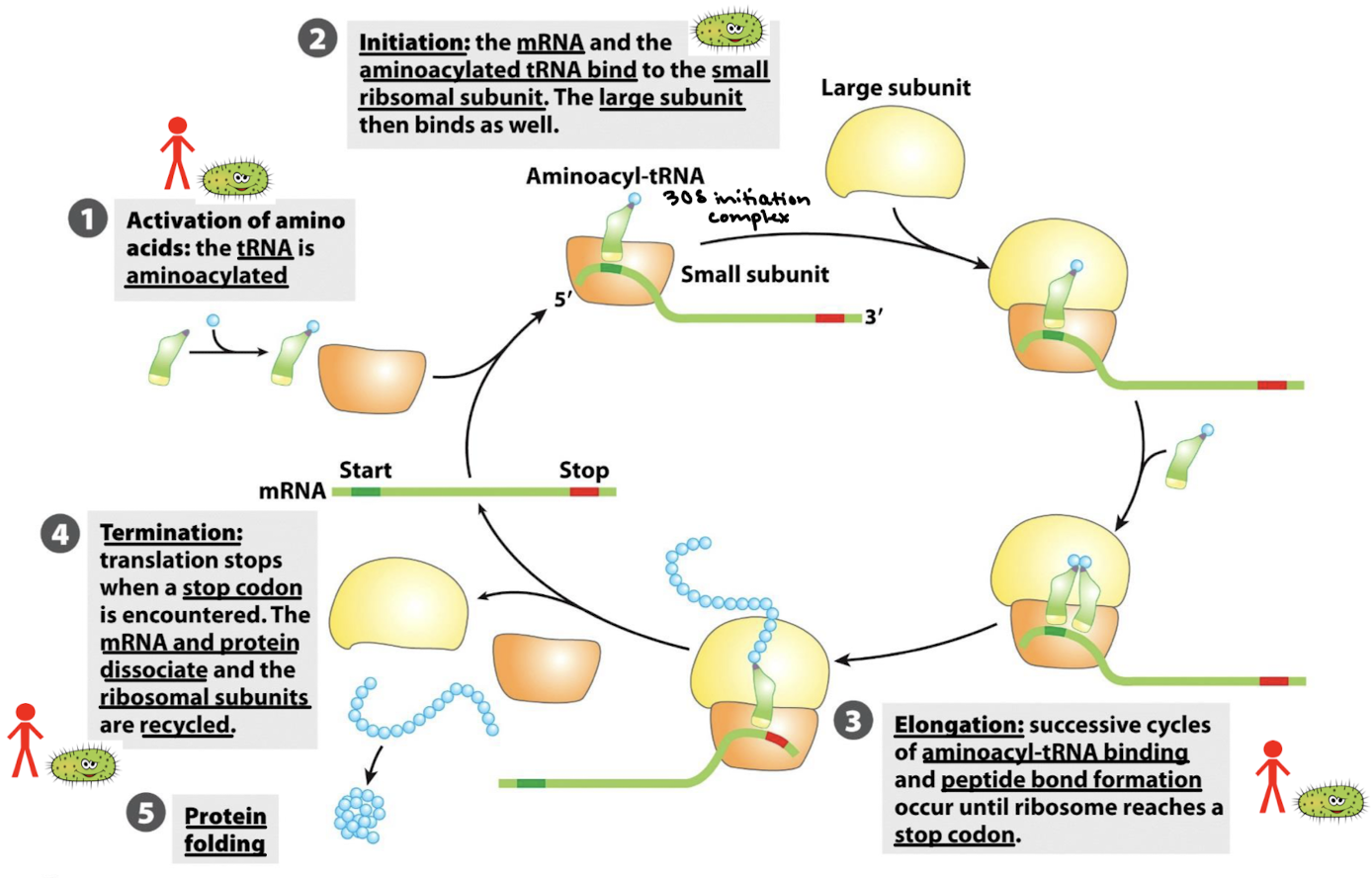
What are the 5 general stages of protein synthesis?
Activation of amino acids: the tRNA is aminoacylated.
Initiation: the mRNA and the aminoacylated tRNA bind to the small ribosomal subunit. The large subunit then binds as well.
Elongation: Successive cycles of amino-acyl-tRNA binding and peptide bond formation occur until ribosome reaches a stop codon.
Termination: translation stops when a stop codon is encountered. The mRNA and protein dissociate and the ribosomal subunits are recycled.
Protein folding.
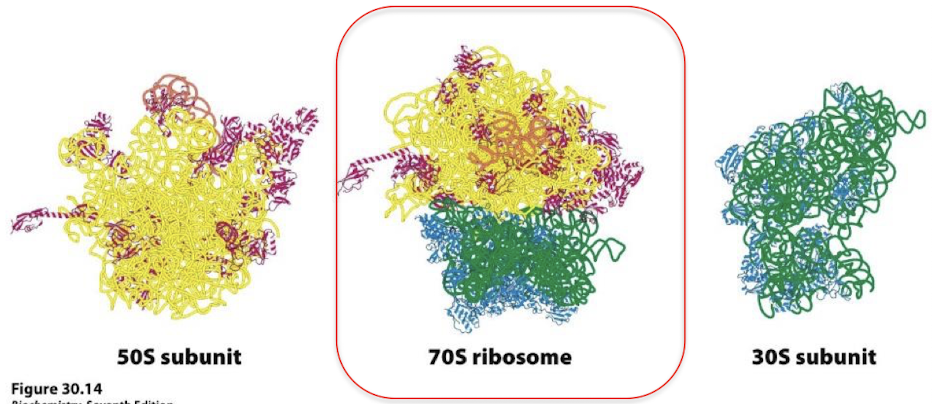
What is the site of protein synthesis? What is the composition?
The ribosome. 65% rRNA and 35% protein (rRNA forms the core).
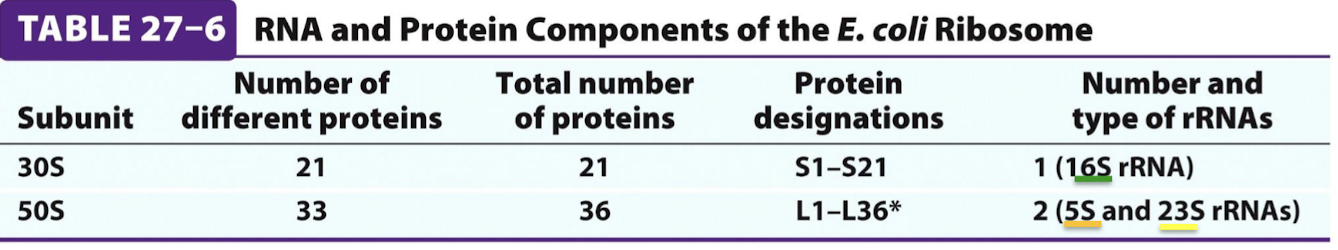
In which subunit are the 5S, 16S, and 23S rRNA of prokaryotes?
The 16S rRNA belongs to the 30S/small subunit and the 5S and 23S rRNAs belong to the 50S/large subunit.
Which rRNAs play a central role in translation? What is that role?
5S, 16S, and 23S rRNA are critical for ribosomal structure & function: rRNA is the actual catalyst for protein synthesis.
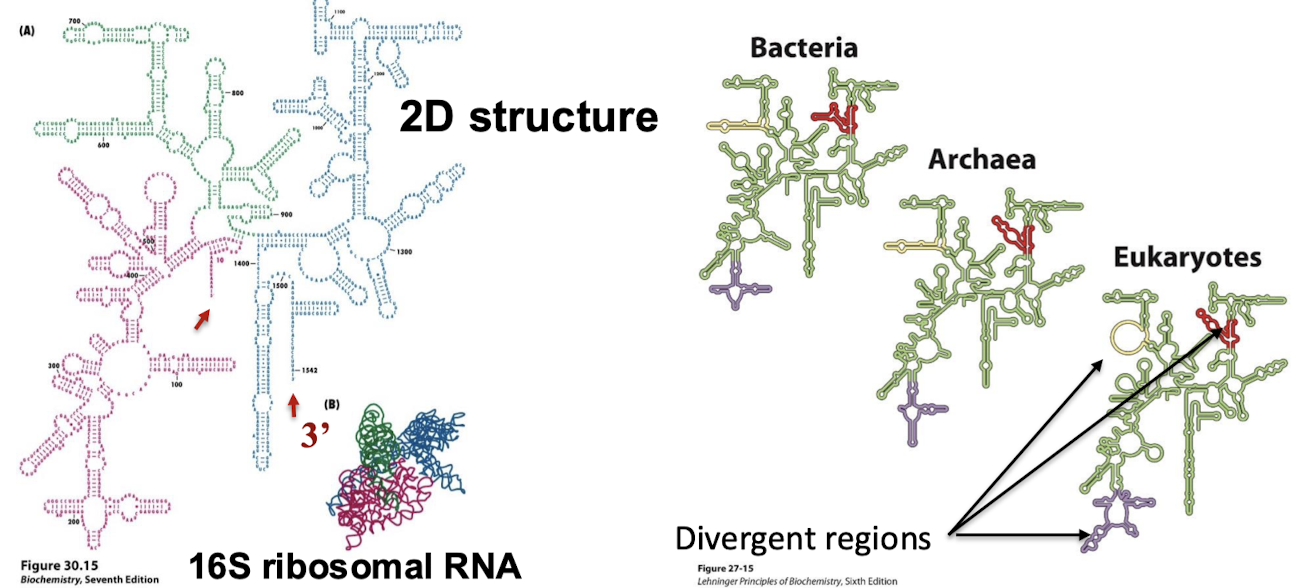
What are the structural characteristics of rRNAs (specifically 5S, 16S, and 23S)?
Specific 3D structure with extensive intrachain base pairs. The shape of rRNAs are highly conserved.
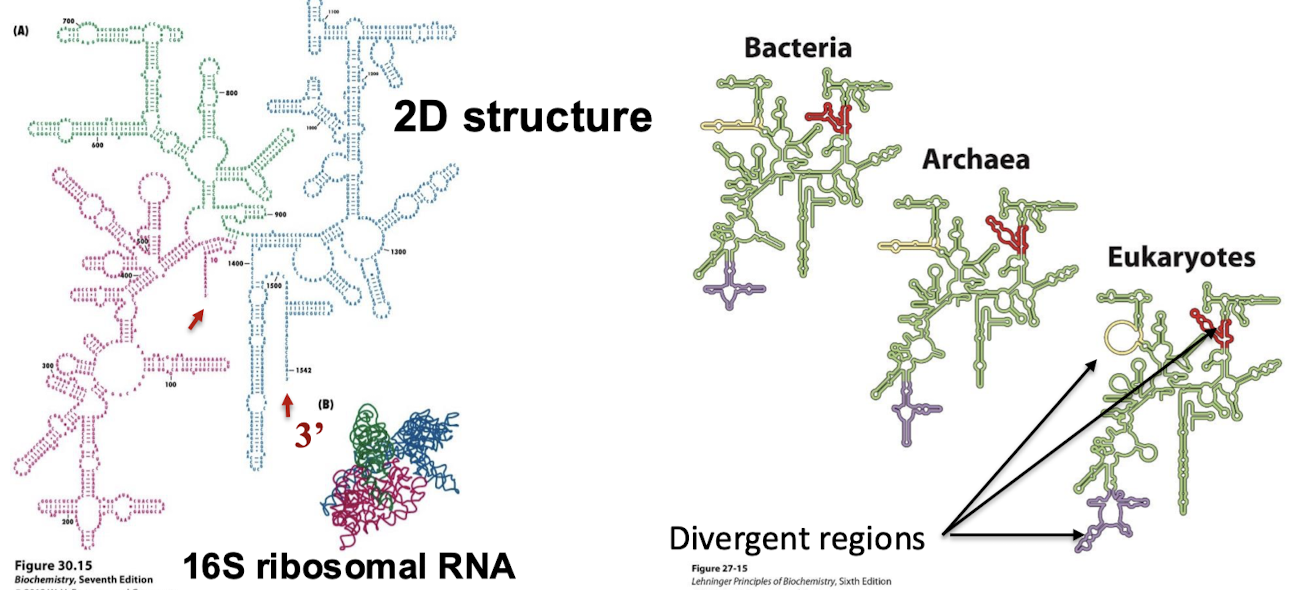
How much larger are eukaryotic ribosomes? What are the different subunits?
30% larger:
Bacterial ribosome: 30S + 50S = 70S
Eukaryotic ribosome: 40S + 60S = 80S
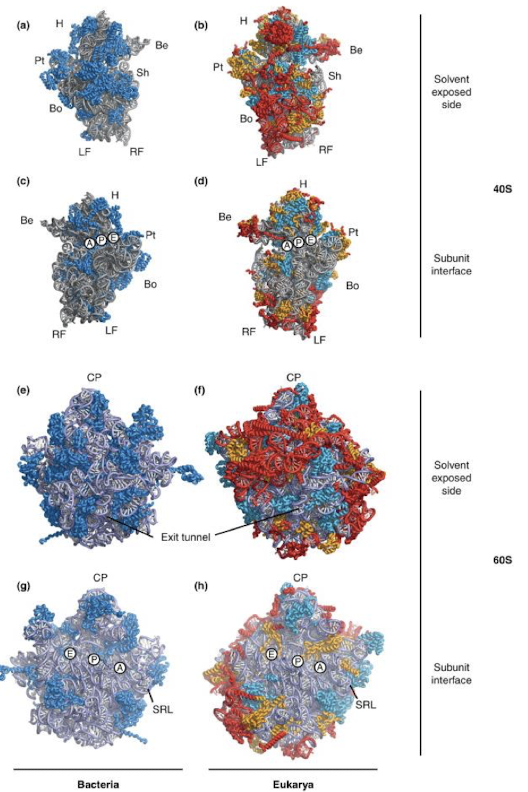
What do the teal, yellow, and red colors represent in the eukaryotic ribosomes?
Teal = universally conserved proteins
Yellow = proteins present in archaea and eukarya
Red = protein and RNA elements exclusively in eukarya
Where is the peptide bond form? Where does the tunnel connect this site to?
The acceptor stems of the tRNA molecules occupying the A site and the P site converge at a site where a peptide bond is formed. A tunnel connects this site to the back of the ribosome.
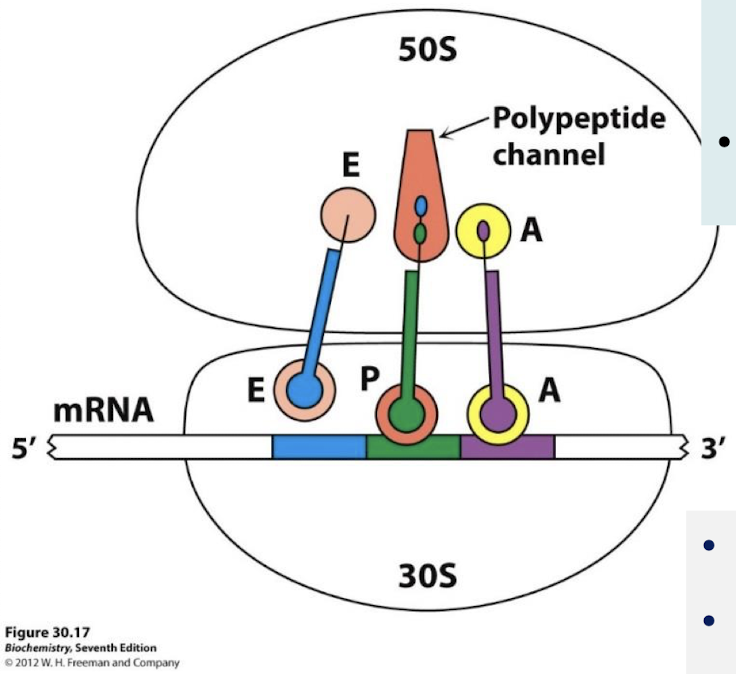
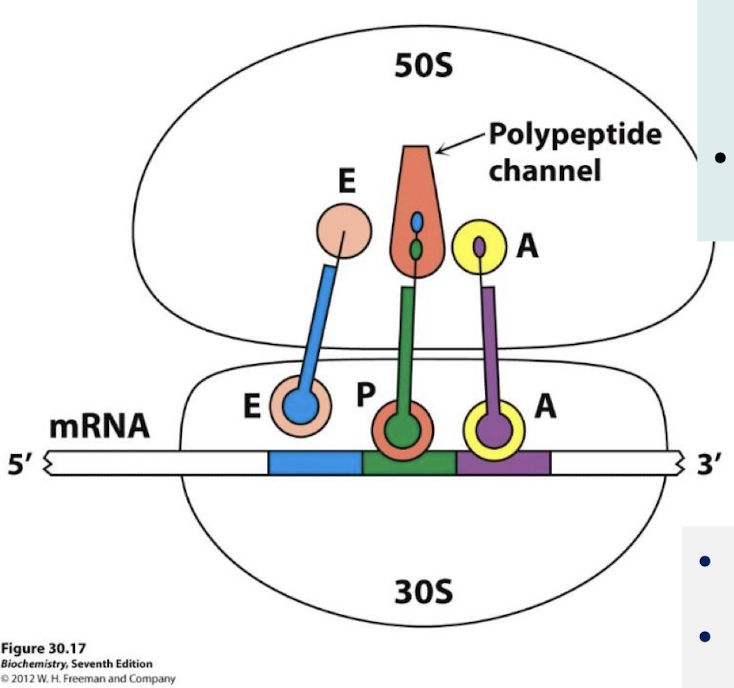
Which subunit holds the mRNA being translated? How do tRNA and mRNA interact?
The small subunit; two tRNA molecules are bound to the mRNA through anticodon-codon base pairs.
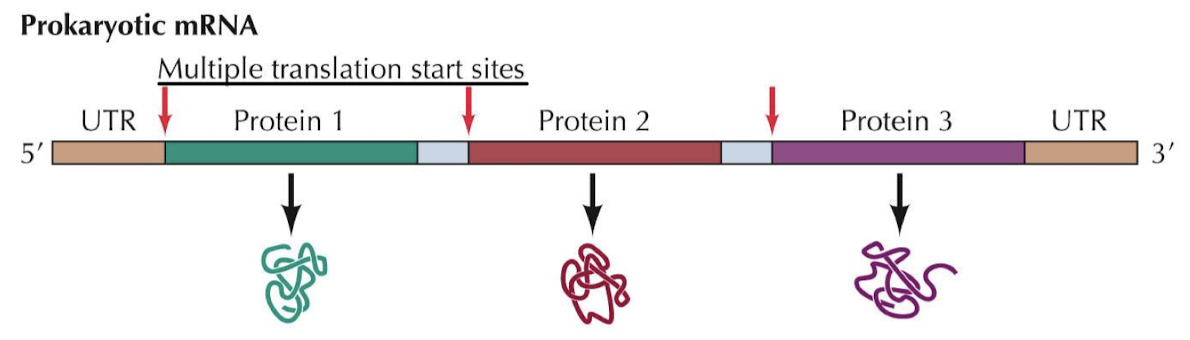
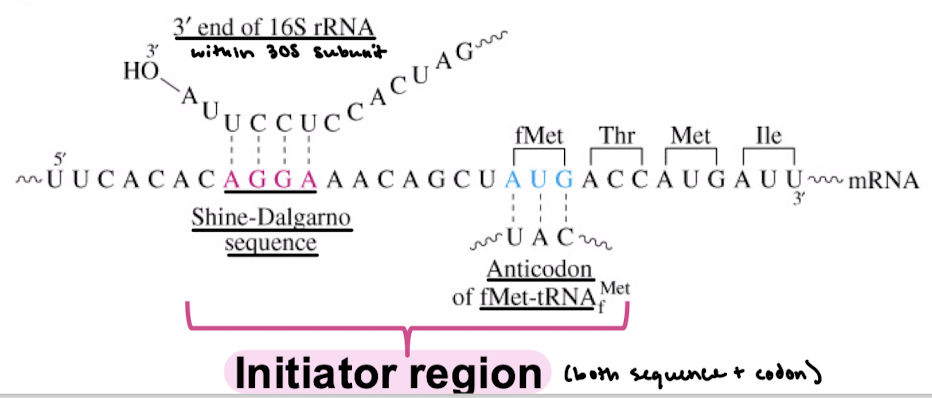
What is the meaning of polycistronic? Which organisms are polycistronic? Do the first three nucleotides of each mRNA serve as the first codon?
Polycistronic refers to an mRNA molecule that encodes two or more different proteins. There are multiple translation start sites on this mRNA. AUG and a Shine-Dalgarno sequence are needed to start.
What rRNA does the Shine-Dalgarno sequence bind? The codon?
The 3’ end of 16S rRNA on the small subunit. The anticodon of fMet-tRNAfMet.
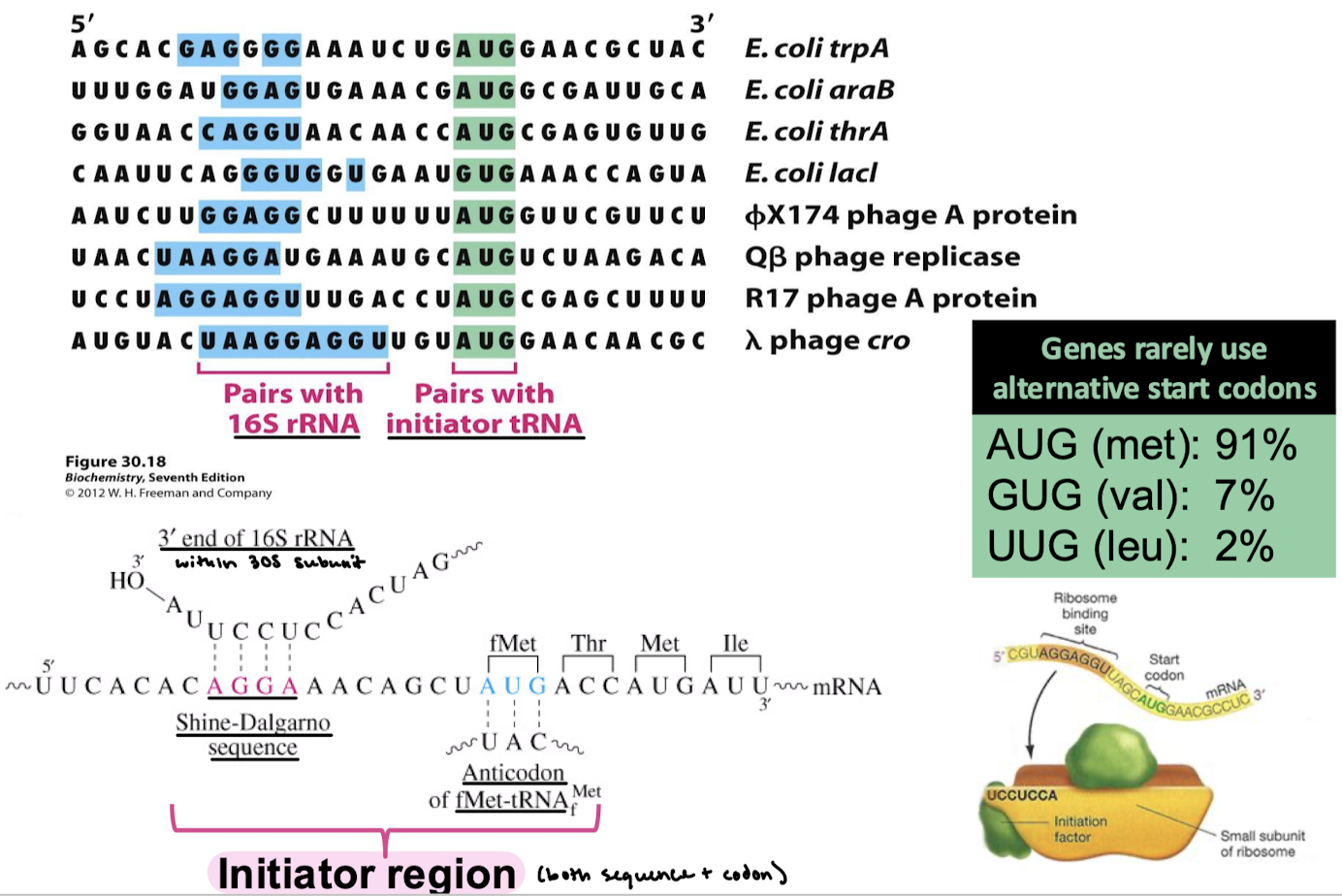
What makes up the initiator region?
Shine-Dalgarno sequence and start codon.
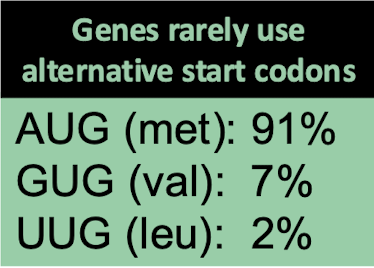
What are altnernative start codons in prokaryotes and which amino acids are associate?
AUG (met): 91%
GUG (val): 7%
UUG (leu): 2%
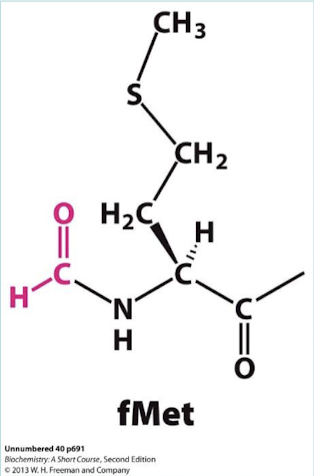
What is the first amino acid in protein synthesis? What is the initiating aminoacyl-tRNA? Is this the same tRNA that incorporates methionine NOT at the start?
N-formyl-methionine (fMet) and N-formyl-methionyl-tRNA (tRNAf). This one differs from the one that inserts methionine in internal positions, tRNAm).
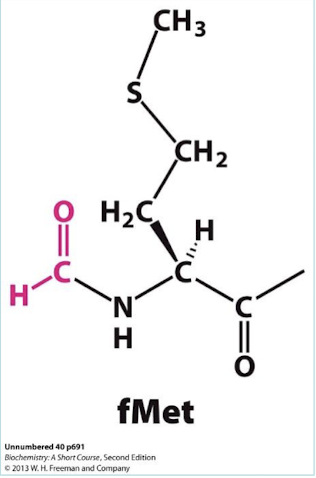
What protein removes fMet from the protein during maturation? what enzyme removes the formyl group?
Methionine aminopeptidase; deformylase.
What are the two functions of fMet? Is it essential?
Interacts with IF2 (efficiency of translational initiation)
Can act as a degradation signal, largely a cotranslational one
It is not strictly essential for protein synthesis and cell viability.
Is the formylation of methionyl-tRNA the same for Met and fMet? What is the enzyme that modifies the methionine attached to tRNAf? What is the activated formyl donor?
The same synthetase activates both tRNAm and tRNAf. The specific transformylase modifies the methionine attached to tRNAf. The activated formyl donor is N10-formyl-tetrahydrofolate.
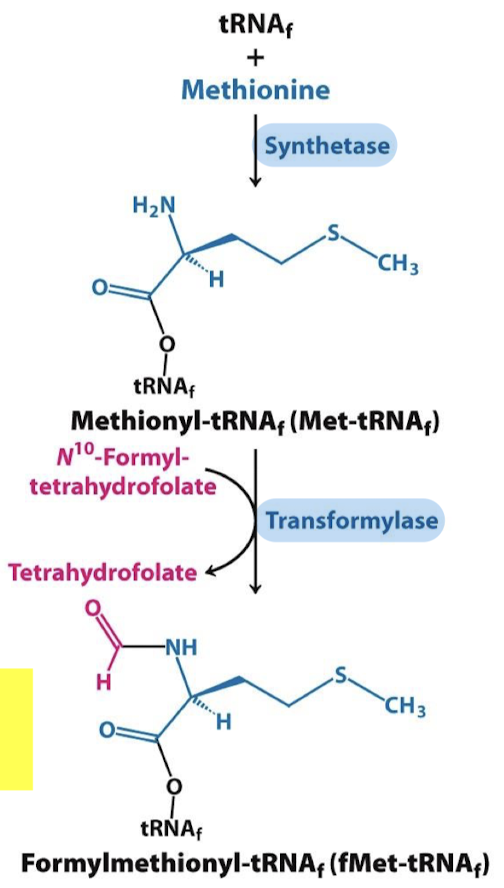
Are free methionine and methionyl-tRNAm substrates for transformylase?
No, but N10-formyl-tetrahydrofolate
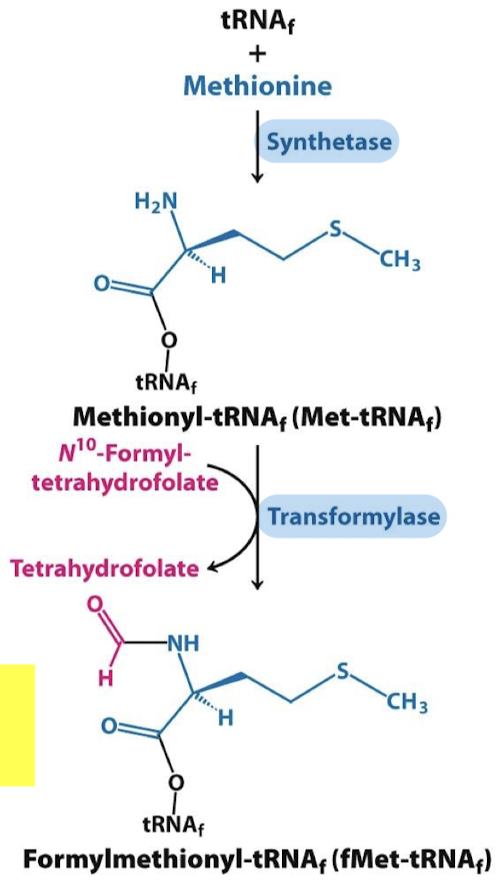
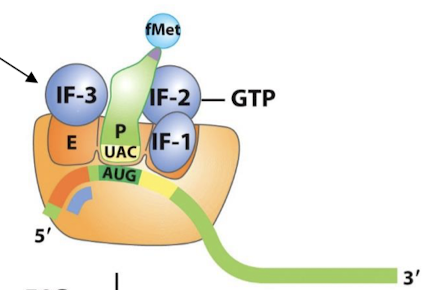
What are the two steps in the formation of the 30S initiation complex in prokaryotes?
The 30S subunit is bound by IF-1 in the A site and IF-3 in the E site, then the mRNA (16S rRNA) at the Shine-Dalgarno sequence.
IF-2—GTP binds the 30S subunit and recruits fMet-tRNAfMet, which base-pairs with the start codon.
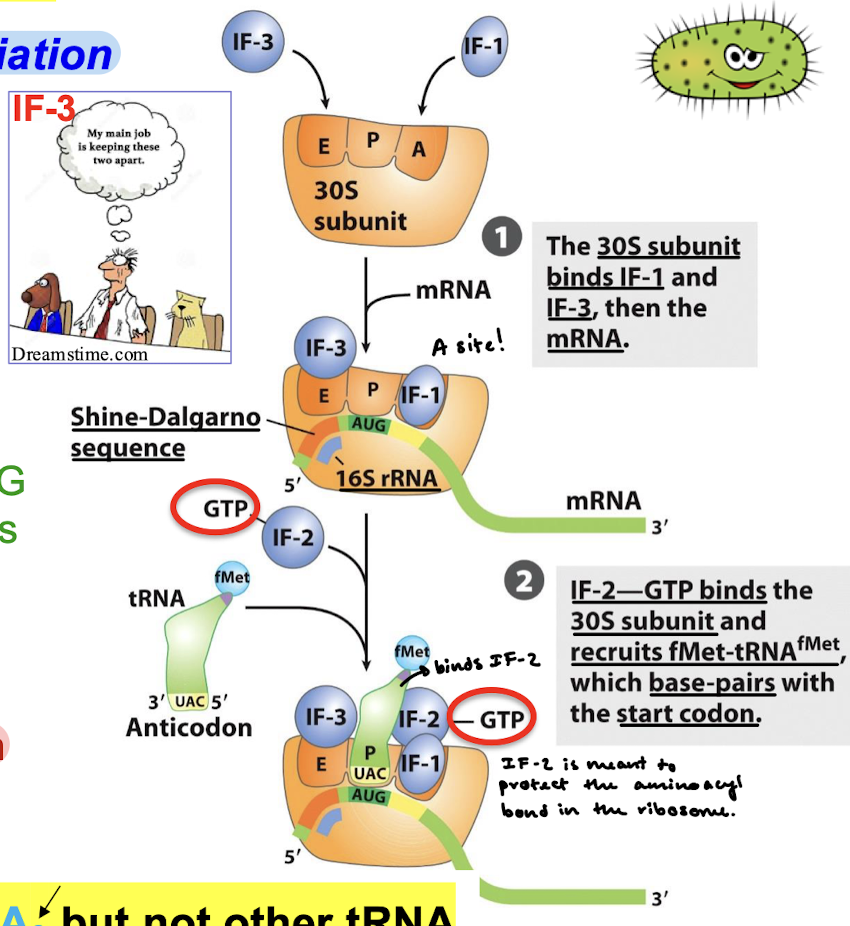
How is the initiating (5’)-AUG codon of mRNA guided to its correct position?
Shine—Dalgarno sequence
IF2 recognizes ________ but not other tRNA
fMet-tRNAf
____ prevents binding of 50S large subunit.
IF-3
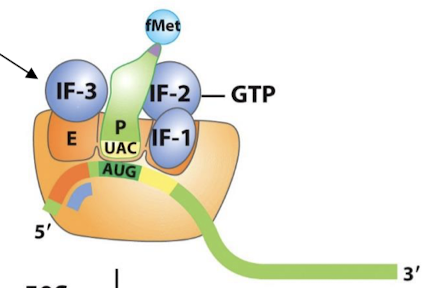
Where does fMet-tRNAf occupy in transcription initiation? What does it bind?
The P site. Binds AUG and establishes the reading frame.
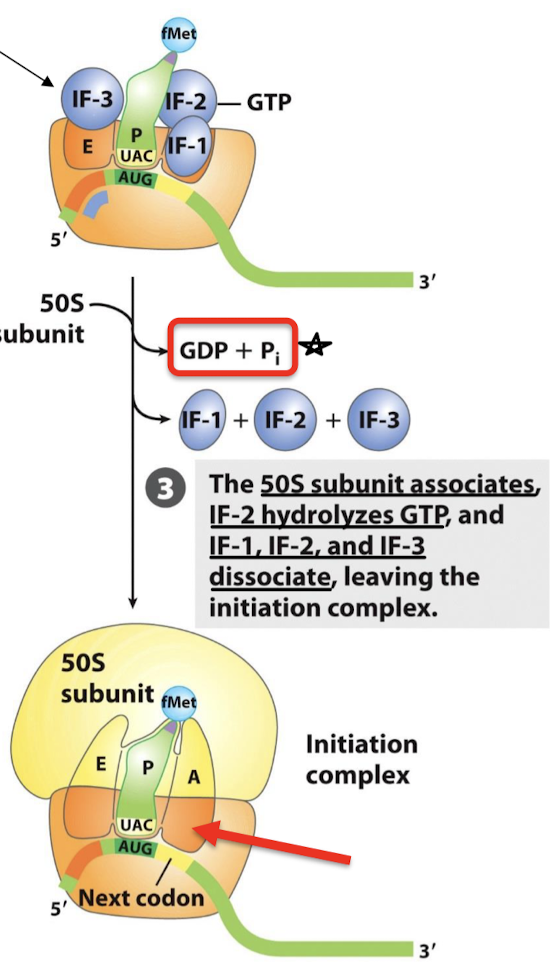
What step is required for the formation of the 70S initiation complex from the 30S initiation complex?
IF-2 hydrolyzes GTP, IF-1, IF-2, and IF-3 dissociate and leave the initiation complex, and the 50S subunit associates.
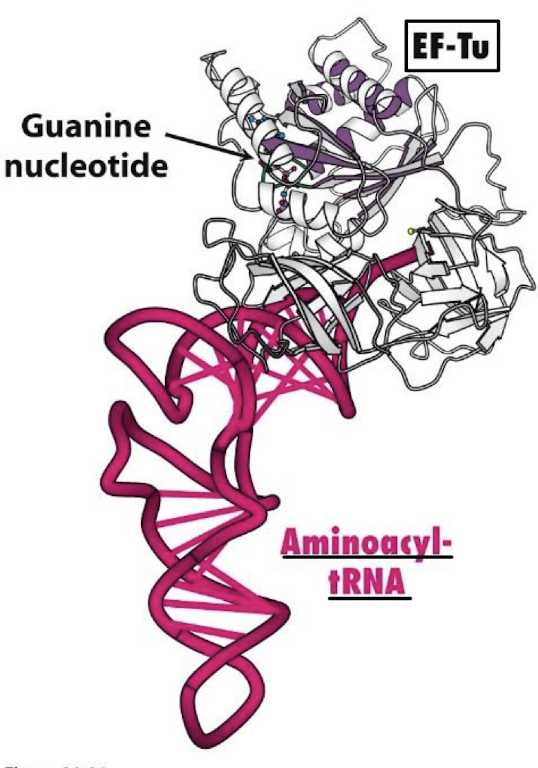
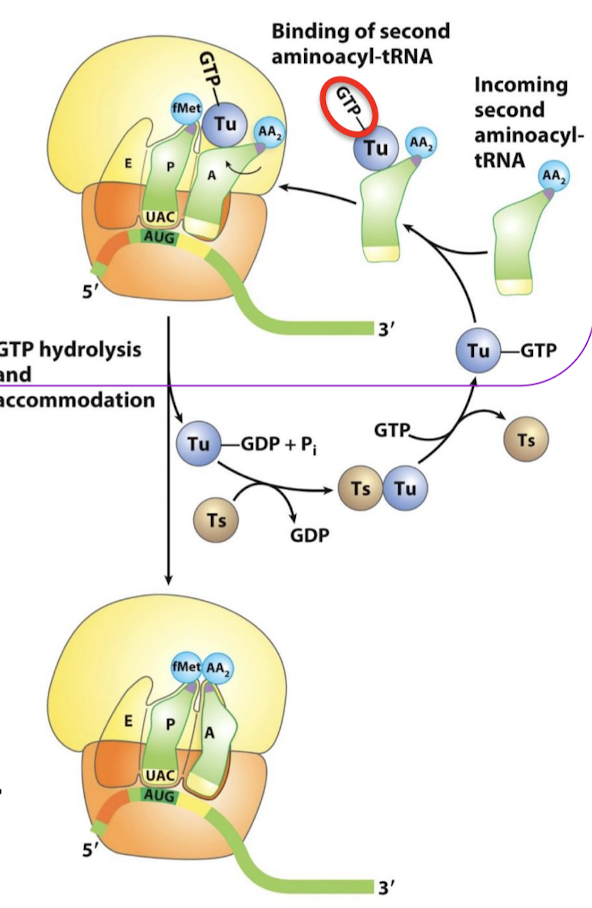
Of what family is EF-Tu? In what form does it bind aminoacyl-tRNA? What is its function? What rRNA does it interact with?
A 43-kd protein, member of the G-protein family. It binds aminoacyl-tRNA only in the GTP form. It delivers the appropriate aminoacyl-tRNA to the A site. It interacts with the 16S rRNA.
The binding of EF-Tu to aminoacyl-tRNA serves what two functions? When is GTP hydrolysis required?
Protects the ester linkage in aminoacyl-tRNA from hydrolysis.
Accuracy of protein synthesis: the GTP in EF-Tu is hydrolyzed to GDP when an appropriate complex between the EF-Tu-aminoacyl-tRNA complex and the ribosome has formed.
What happens if the anticodon is not properly paired with the codon in the EF-Tu-aminoacyl-tRNA complex? Why is this beneficial?
Hydrolysis does not take place, and the aminoacyl-tRNA is not transferred to the ribosome. This mechanism allows the free energy of GTP hydrolysis to contribute to the fidelity of protein synthesis.
With which aminoacyl-tRNA does EF-Tu-GTP NOT bind?
Does NOT interact with fMet-tRNAf. Met-tRNAm does bind to EF-Tu like all other aminoacyl-tRNAs.
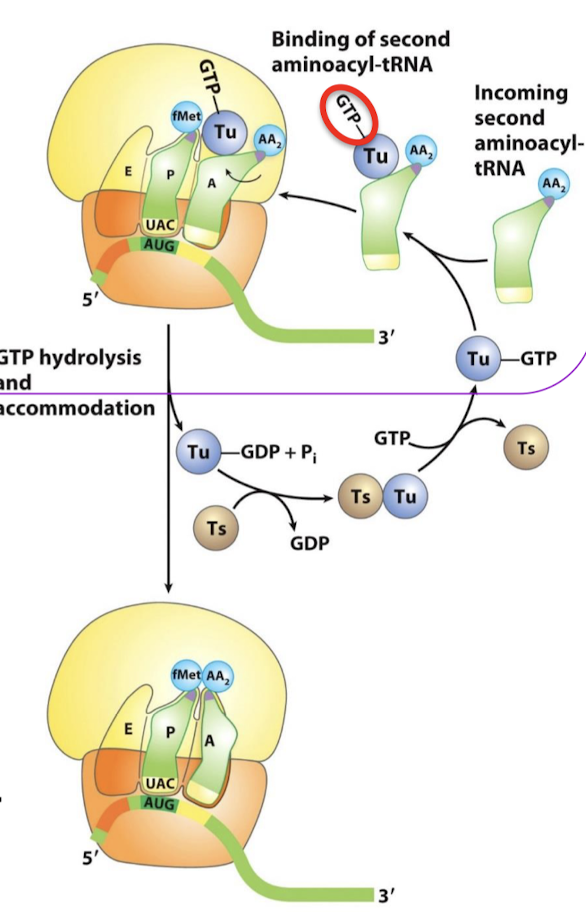
What is the role of EF-Tu-GDP?
It dissociates from the aminoacyl-tRNA, allowing the tRNA to be fully accommodated into the A site.
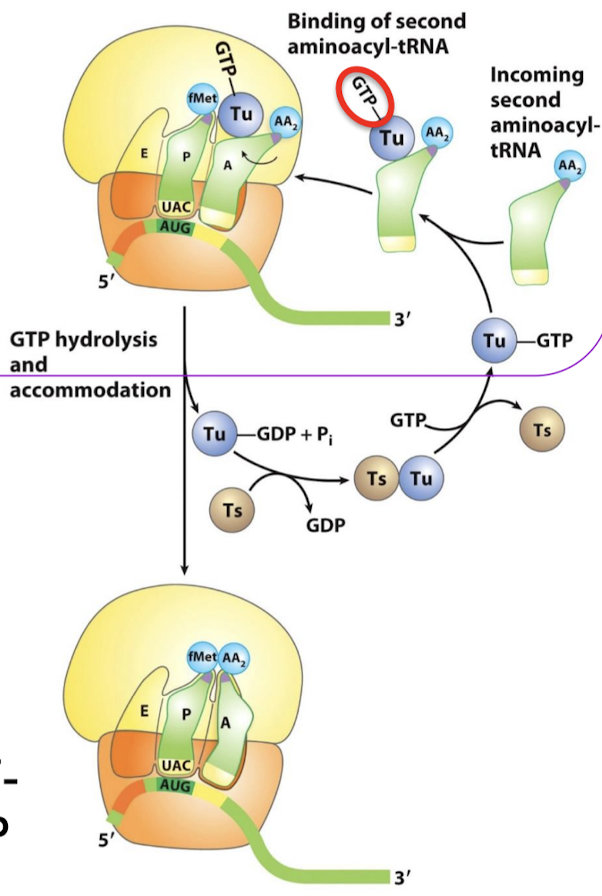
What is the role of EF-Ts?
EF-Ts act as a GEF (guanine nucleotide exchange factor) for EF-Tu. It promotes the release of GDP to allow the rebinding of a GTP.
Which rRNA ribozyme functions as the peptidyl transferase? What interacts in the peptidyl transferase center?
23S rRNA for peptide bond formation. The aminoacetylated ends of both A- and P-site tRNA interact at the PTC of the 50S subunit.
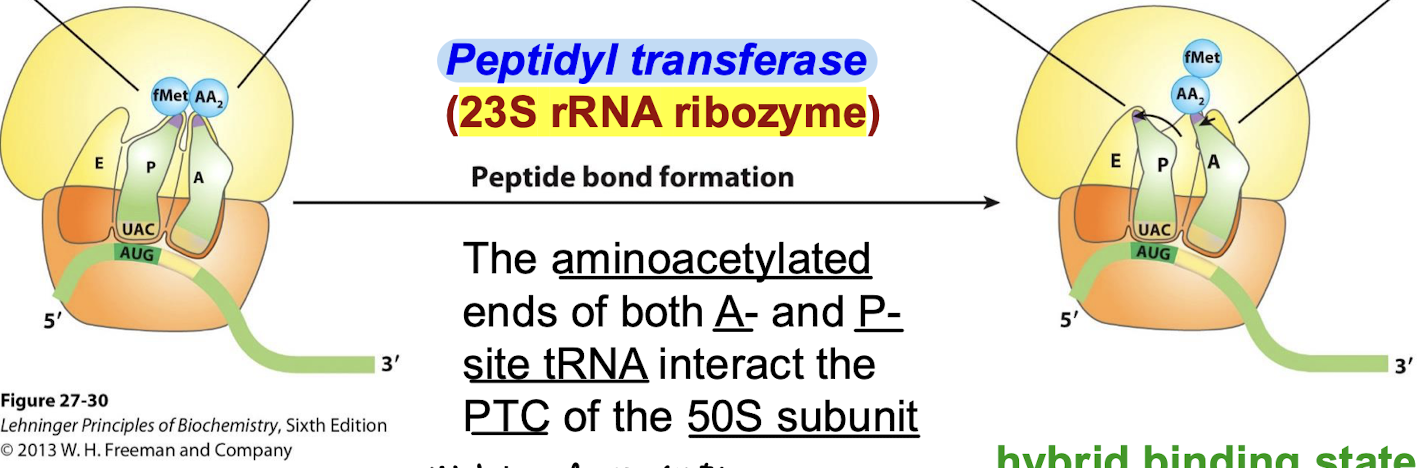
What is the role of the PTC?
To align the tRNA-linked substrates so that the reaction is facilitated.
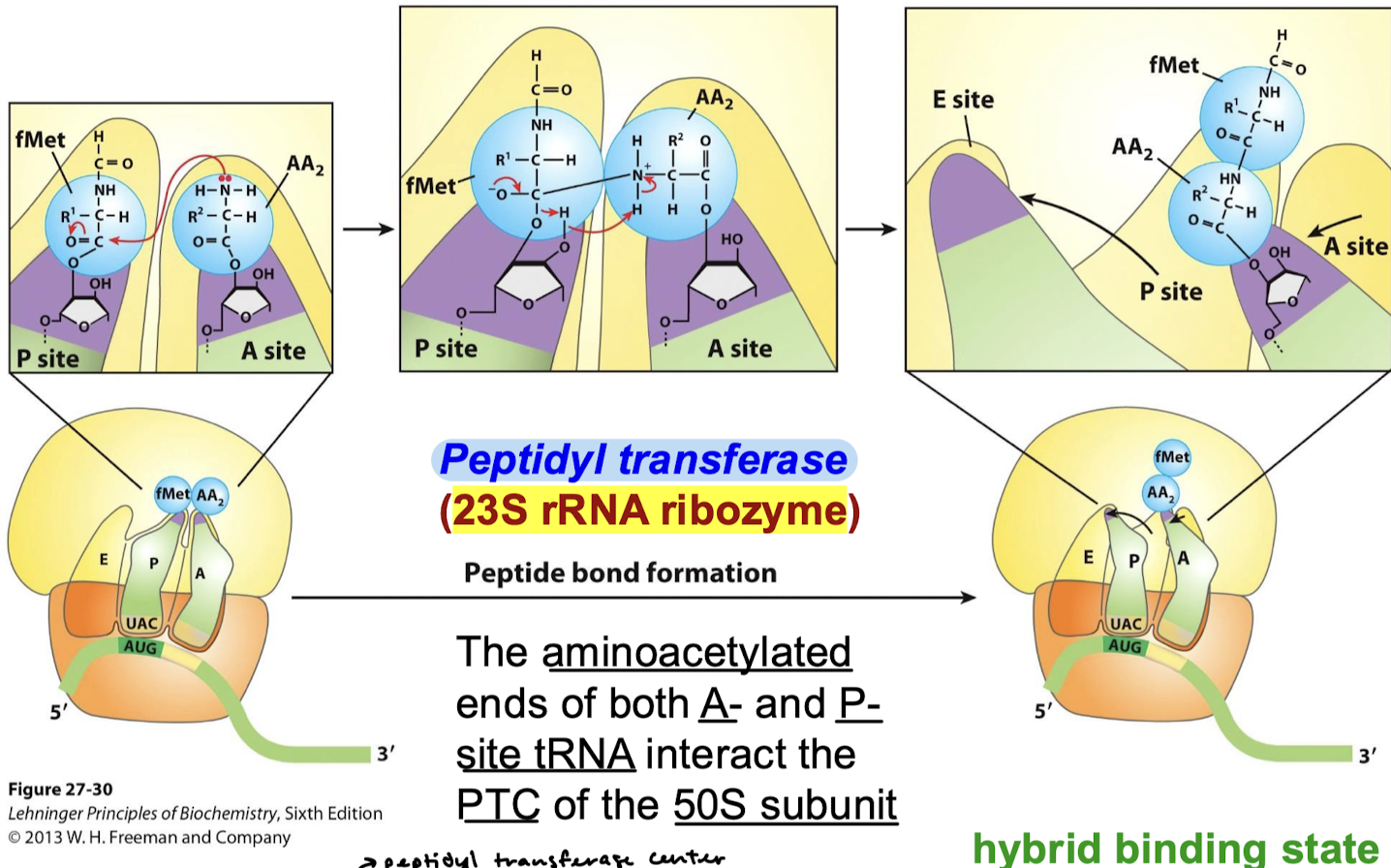
Describe the reaction that occurs in the PTC.
Peptide bond formation: AA2’s amino group nitrogen nucleophilically attacks the carbonyl carbon involved in the aminoacyl ester linkage, forming a bond between fMet and AA2. Various electron movements result in the breakage of the ester linkage between fMet and the P-site tRNA acceptor stem.
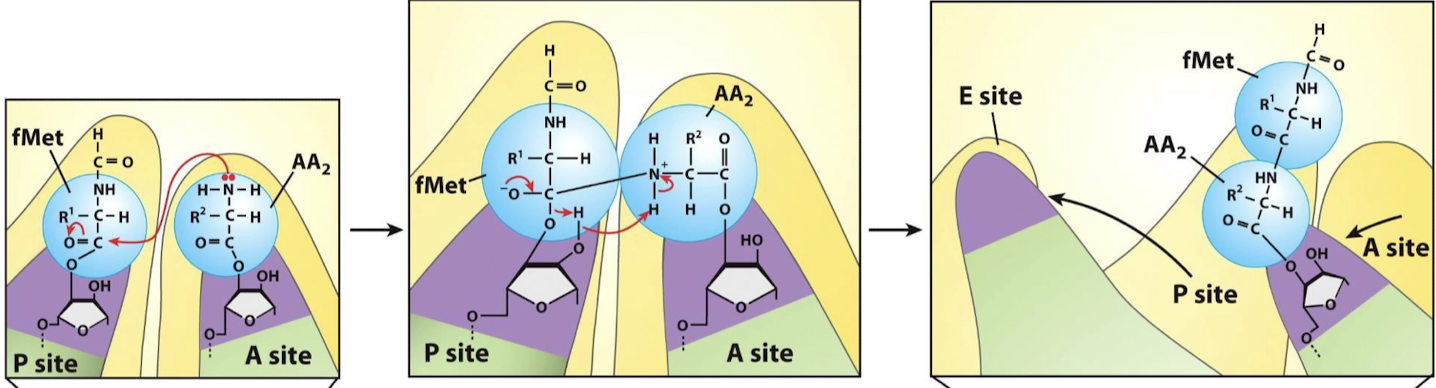
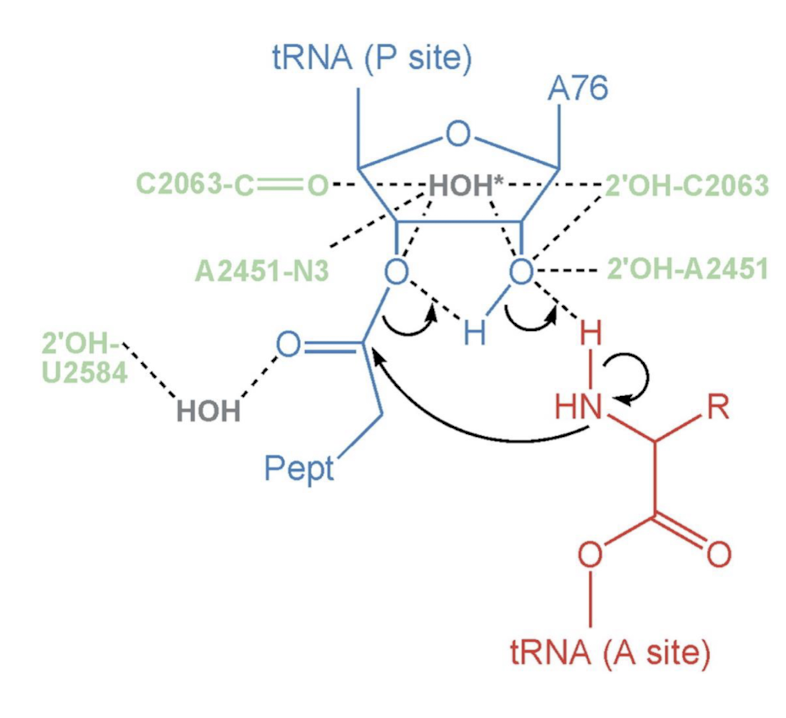
How does 23S rRNA catalyze peptide bond formation? Describe the formation reaction.
The rRNA stabilizes the deprotonated alpha amino group (NH2 instead of the NH3+ common at neutral pH). The orientation facilitates nucleophilic attack of alpha amine to ester linkage, forming a peptide bond with the carboxyl group of the previous amino acid.
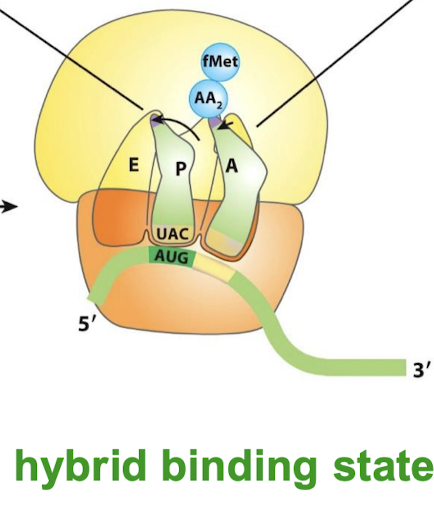
Describe the hybrid binding state:
Uncharged tRNA → 3’ and 5’ ends in the E site.
Peptidyl tRNA —> 3’ and 5’ shift to the P site.
The anticodons remain in the P and A sites.
What is the state called that occurs upon peptide bond formation?
Hybrid binding state
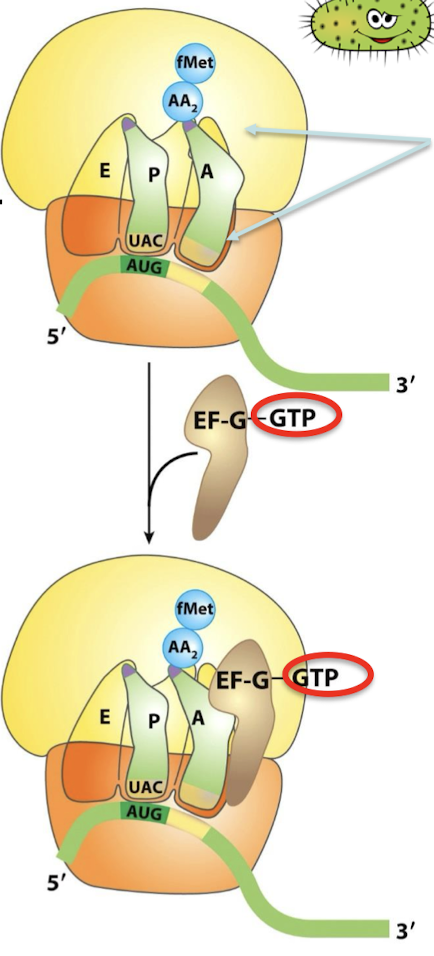
What is the function of EF-G? What form does it bind? Where? With what rRNA and on what subunit?
EF-G is the translocase. In the GTP form, EF-G binds to the ribosome near the A site, interacting with the 23S rRNA of the 50S subunit.
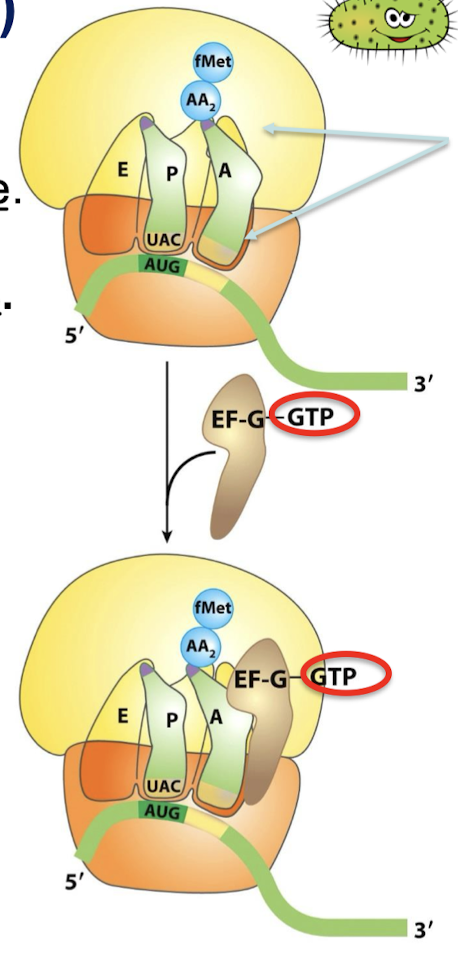
What stimulates GTP hydrolysis in EF-G-GTP?
Binding of EF-G to the ribosome.
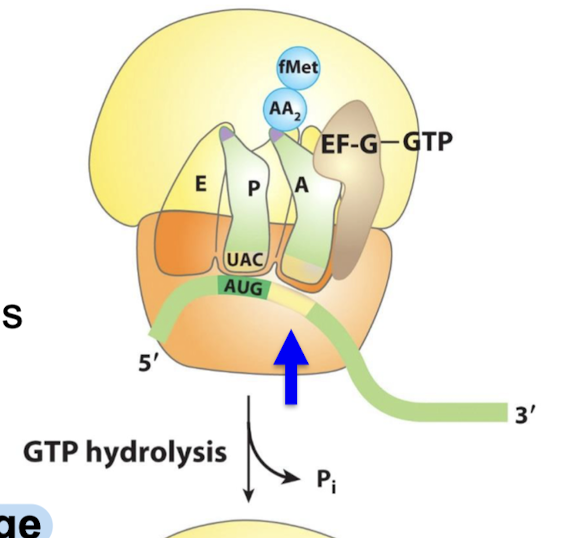
What results form EF-G GTP hydrolysis?
EF-G undergoes a conformational change that displaces the peptidyl-tRNA in the A site to the P site → carries the mRNA and the acetylated tRNA with it.
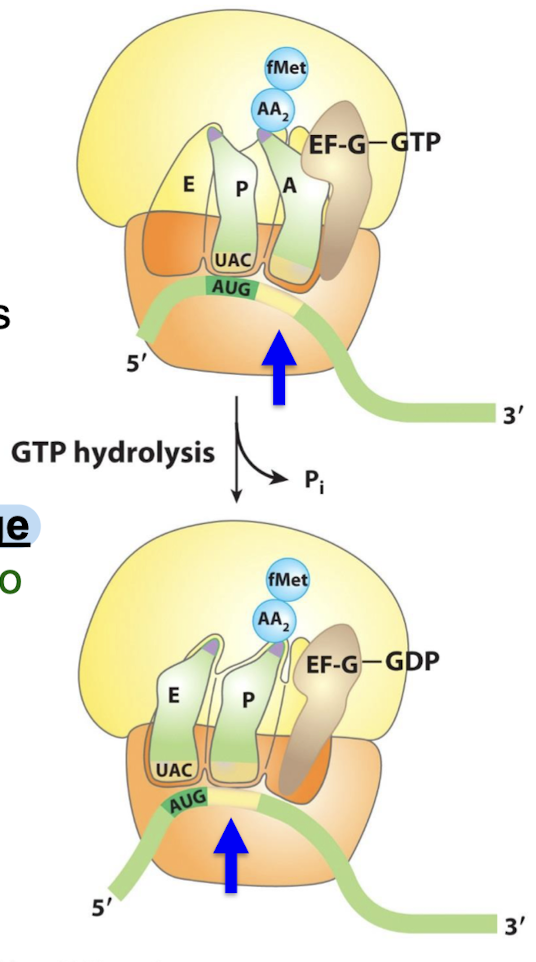
Where does the peptide chain remain as translocation occurs?
The peptide chain remains in the P site on the 50S subunit throughout this cycle, growing into the exit tunnel.
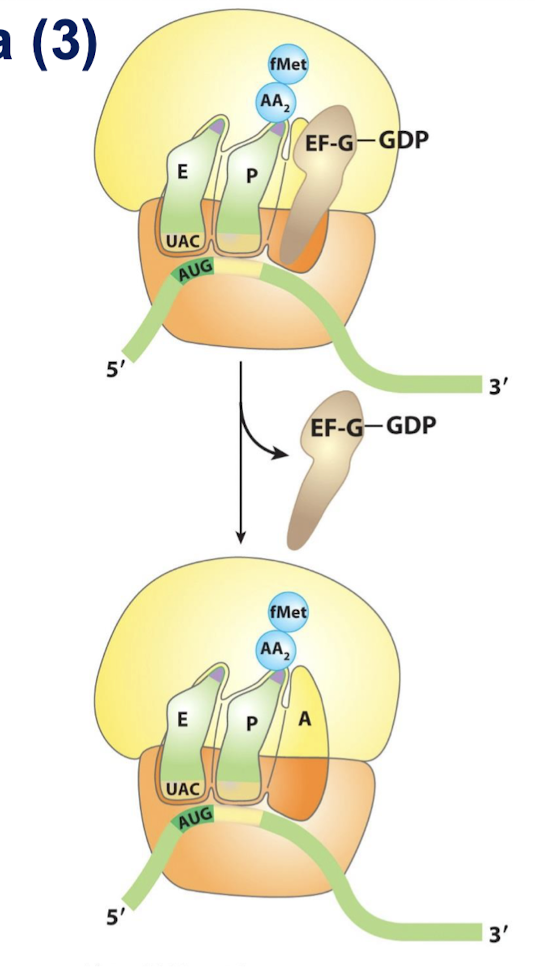
What direction does mRNA translation take place?
5’ to 3’
What is the final phase of translation? When does this process occur?
The elongation process continues until a stop codon appears in the A site, such as UAA, UGA, and UAG.
What recognizes the STOP codons?
Stop codons are recognized by proteins called release factors (RFs). There are NO tRNAs that recognize the STOP codons.
Which release factors recognize what stop codons? What is the function of the one that doesn’t recognize stop codons?
RF1: UAA or UAG
RF2: UAA or UGA
RF3: A GTPase, mediates interactions between RF1 or RF2 and the ribosome.
Where do release factors bind? Where does RF interact with the PTC?
The RF binds to the ribosome, the proteins unfold to bridge the gap between the stop codon on the mRNA and the peptidyl transferase center (PTC) on the 50S subunit. The RF interacts with the PTC with a loop containing a highly conserved Gly-Gly-Gln (GCQ) sequence, with the Gln methylated.
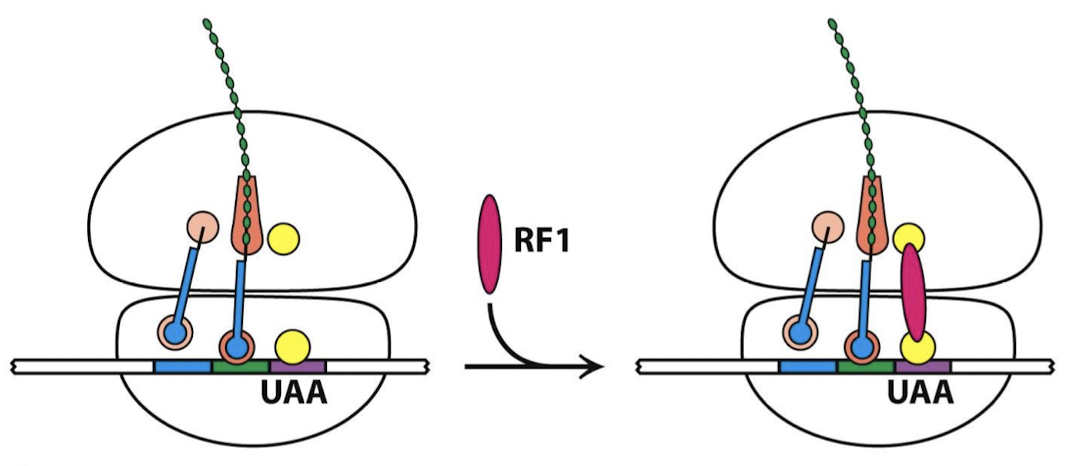
What are the two steps in termination of protein synthesis?
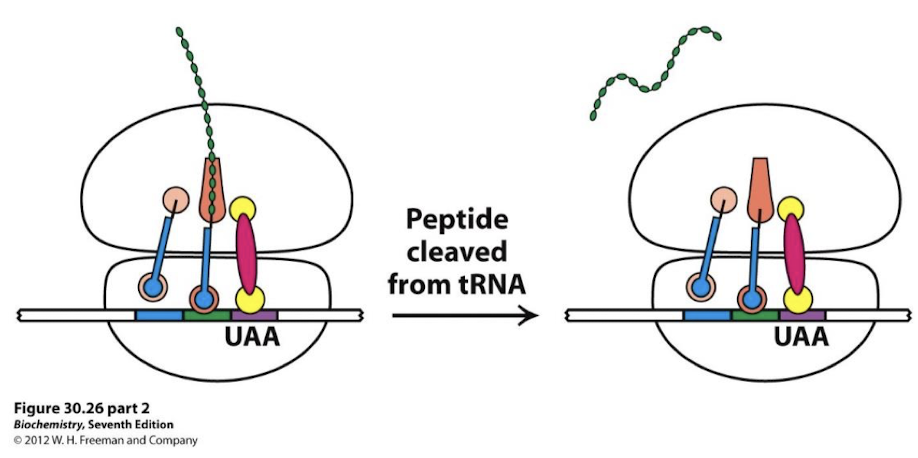
Binding of release factors
Hydrolysis of the peptidyl-tRNA.
What facilitates the hydrolysis of the peptidyl-tRNA?
Modified Gln, along with the peptidyl transferase, facilitates the attack of a water molecule on the ester linkage between the polypeptide chain and tRNA in the P site, releasing the complete protein.
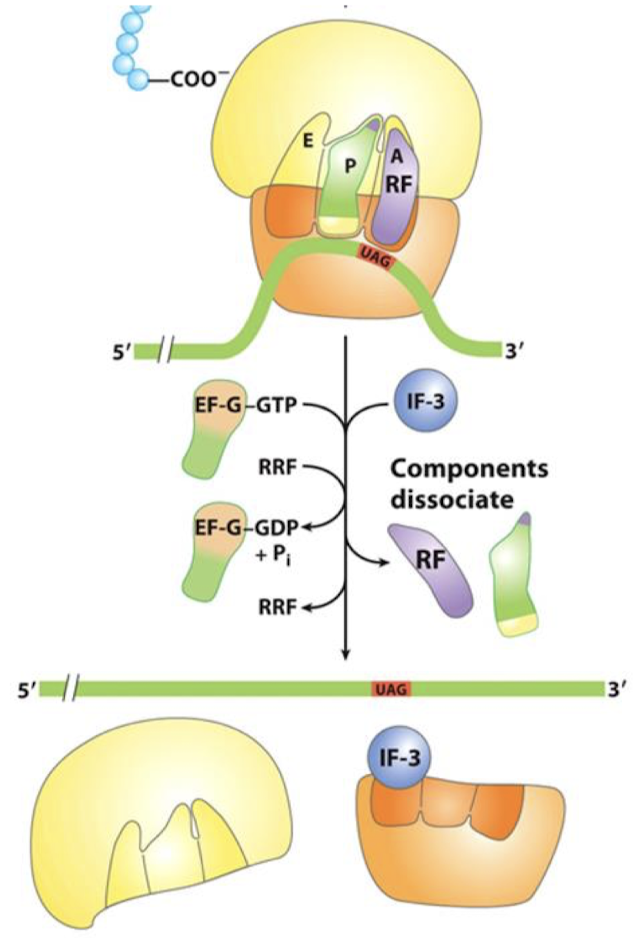
What causes dissociation of the ribosome, mRNA, and attached tRNA?
EF-G and ribosome release factor (RRF) which is facilitated by GTP hydrolysis of EF-G-GTP. IF-3 binds the E-site of small subunit which causes components to dissociate.
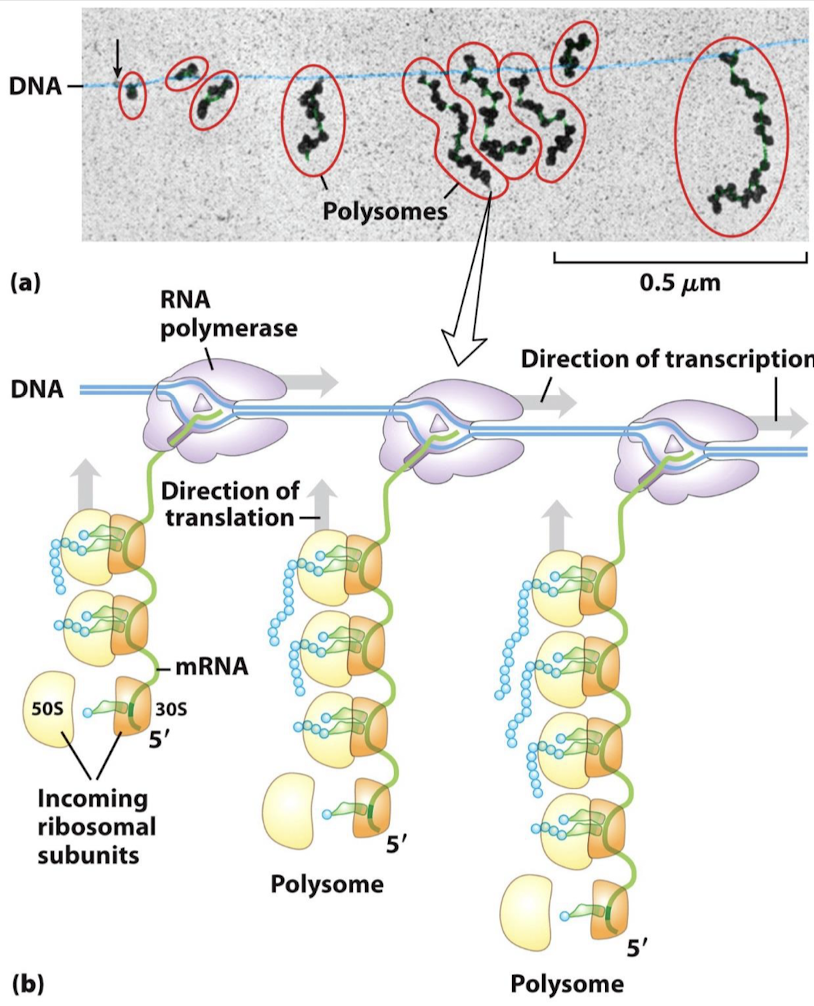
What are polysomes?
An mRNA being transcribed with ribosome translating simultaneously.