6.2- Patterns of Inheritance
1/58
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
59 Terms
gene
a sequence of bases on a DNA molecule that codes for a protein
allele
different versions of the same gene that code for variants of a characteristic
genotype
organism’s genetic makeup→ combination of alleles
phenotype
the characteristics expressed by the genotype
dominant
an allele that is always expressed it the phenotype→ only 1 copy present to express phenotype
recessive
allele that is only expressed when homozygous in genotype
locus
specific position of gene on chromosome
homozygous
2 identical alleles for a trait
heterozygous
two different alleles for a trait
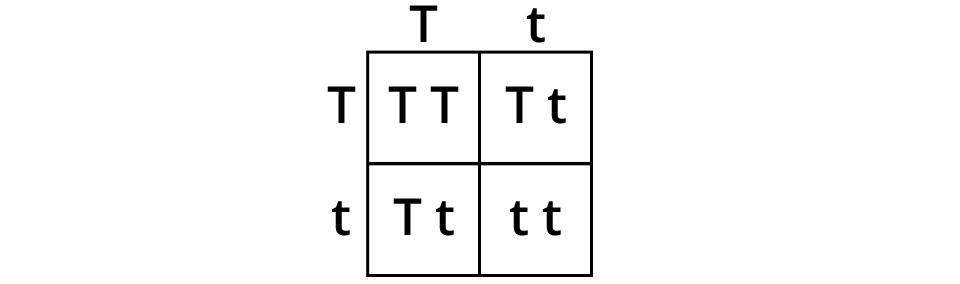
ratio for heterozygous monohybrid cross
genotypic ratio→ 1:2:1
phenotypic ratio→ 3:1
codominance
when two different alleles are equally expresses in an organisms phenotype→ phenotype is a mixture of both alleles rather than just one
uppercase letter used to signify gene and superscript letters used to indicate alleles e.g. CR and CW for red red and white colour
multiple alleles
genes that exist in more than two allelic forms
organism can only have two alleles of a specific gene at once
e.g. blood groups:
IA, IB, IO alleles exist
inheritance of sex in humans
23rd pair of chromosomes are sex chromosomes:
X chromosome→ found in both male and females
Y chromosome→ only found in males
Female body cells→ XX so gametes always contain X
Male body cells→ XY so gametes contain either X or Y
sex linkage
genes located on the X or Y chromosomes→ sex linked
X chromosome carries most of the genes→ larger
recessive alleles on X chromosome appear in phenotype more often in males- no corresponding allele on Y to mask them
haemophilia
X-linked recessive disorder→ caused by a defective gene on the X chromosome
allele alters DNA sequence coding for a blood clotting protein→ faulty allele= reduced blood clotting so excessive bleeding
inheritance of haemophilia
mainly affects males→ no second X chromosome to mask faulty allele
Doesn’t often affect females→ 2 alleles must be inherited for expression
always inherited from mother in males→ males always inherit X chromosome from their mother
mainly inherited from carrier mothers
affected fathers can only pass it on to daughters→ only daughters inherit X chromosomes from their father
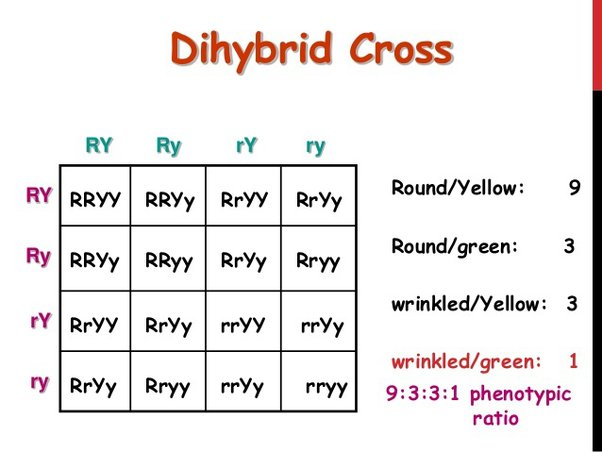
dihybrid cross
shows the simultaneous inheritance of two genes controlling separate characteristics
helps to:
determine if genes are linked
locate genes on specific chromosomes
calculate expected phenotypic ratios in subsequent generations
ratio for heterozygous dihybrid cross
9:3:3:1
autosome
chromosomes that do not determine the sex of an organism→ first 22 chromosome pairs
autosomal linkage
when genes are linked on the same autosome
inherited together in offspring rather than assorting independently
results of autosomal linkage
non-random association alleles at different loci
phenotypic ratios are different from those expected
parental allele combination preserved across generations
how does crossing over affect autosomal linkage
crossing over can separate linked genes
when genes are linked, fewer offspring have different combinations of alleles from their parents due to crossing over (recombinant)
less genetic variation introduced from crossing over when genes are linked
the closer the gene, the more likely they will be inherited together
calculating recombination frequency
50%= no linkage between genes
less than 50% = some degree of autosomal linkage
lower frequency= closer genes are located to each other on chromosome- less chance of being separated
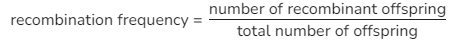
epistasis
an interaction between genes where one gene affects/ masks the expression of another gene
hypostatic gene
gene whose expression is affected by another gene
epistatic gene
gene whose allele affects the expression of the hypostatic gene
phenotypic ratio in epistasis
9:4:3
recessive epistasis
occurs when epistatic gene must be homozygous recessive to block expression of hypostatic gene
dominant epistasis
occurs when epistatic gene is dominant and actively modifies or blocks the expression of hypostatic gene
what is chi squared test used for
assessing whether outcomes of a genetic cross are significantly different from outcomes predicted by specific inheritance pattern.
criteria for chi squared test
large sample size
discrete data categories
using raw counts
comparison of experimental and theoretical results
steps in chi squared test
null hypothesis→ no significant difference between observed and expected, any difference due to chance
alt hypothesis→ significant difference between observed and expected results, difference due to factor other than chance
predict expected phenotypic ratios among offspring
conduct crosses and record observed ratios
calculate chi squared statistic
compare to critical value at chosen probability level→ if χ2 is greater than critical value, reject H0
degrees of freedom
number of phenotypes-1
factors affecting evolution
mutation
gene flow
genetic drift
natural selection
sexual selection
gene flow
the transfer of alleles within the population
genetic drift
random changes in allele frequencies within a population’s gene pool due to chance events
does not occur due to natural selection
can accelerate development of new species in small isolated populations
categories of factors limiting population size
density dependent factors→ depend on size of population e.g. competition, predation, disease
density independent factors→ impact pop. regardless of size e.g. natural disasters, climate change
bottleneck effect
when pop. size reduces suddenly and drastically and reduction lasts for at least 1 generation
can lead to reduced gene pool and genetic diversity→ causes issues related to inbreeding and reduced fertility
may also allow beneficial mutation to become more prevalent
founder effect
when a small group splits from a larger population and small new population is established by this small number of individuals
can lead to reduced gene pool and decreased genetic diversity
rare alleles may become more common in new population
how does variation drive selection
generates range of phenotypes within population, enhancing likelihood that some individuals have alleles for advantageous traits
individuals with beneficial traits are more likely to survive and reproduce under changing conditions
natural selection occurs
types of selection
directional
stabilising
disruptive
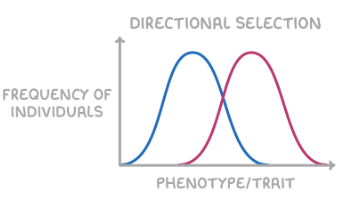
directional selection
selects for one extreme phenotype over others
increases allele frequency for one extreme phenotype
shifts curve in direction of favoured extreme
e.g. antibiotic resistance in bacteria
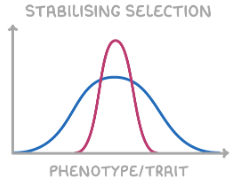
stabilising selection
selects for the average phenotype and selects against extreme phenotypes
increases allele frequency for average phenotype, decreases frequency for extremes
narrows curve
e.g. human birth weights
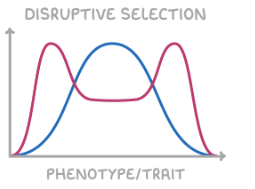
disruptive selection
increases allele frequency for multiple extreme phenotypes, decreases allele frequency for intermediates
curve shifts into multiple peaks either side of average peak
e.g. bird beaks adapting to become larger and smaller when there are different food sources
hardy weinberg principle
helps calculate allele frequency for a gene in a population
assumptions made in hardy weinberg principle
no mutations
no migration into or out of population
mating is random
large population size
no natural selection pressures
variables in hardy weinberg
p → frequency of dominant allele
q → frequency of recessive allele
p2 → frequency of homozygous dominant
2pq→ frequency of heterozygous
q2 → frequency of homozygous recessive
hardy weinberg principle equations
p+q=1
p2+2pq+q2 =1
reproductive isolation
when population cannot interbreed successfully to produce fertile offspring
results in genetic isolation
prezygotic reproductive barriers
prevent fertilisation and formation of a zygote
e.g. habitat isolation, variations in mating rituals
postzygotic reproductive barriers
often a result of hybridisation between different species
produce infertile offspring, reducing reproductive potential
allopatric speciation
some members of population are geographically isolated by a physical barrier e.g. mountain, river, sea
geographical separation exposes distinct parts of population to different environmental pressures
prezygotic reproductive barriers= reproductive isolation
reproductive isolation prevents gene flow and physical separation= genetic divergence
causes populations to evolve separately and form separate species
sympatric speciation
takes place within same geographical location
ecological or behavioural separation mechanisms e.g. different habitat preference lead to groups becoming reproductively isolated
reproductive isolation prevents gene flow and leads to genetic divergence
causes populations to evolve separately and form separate species
what does speciation occur due to
reproductive isolation of populations
genetic divergence→ natural selection and genetic drift
adaptive radiation
organisms diversify rapidly from ancestral species into wide array of new forms→ each adapted to specific ecological niche
more likely to occur when change in environment makes new resources available
artificial selection
when humans breed organisms selectively for specific genetic traits and determine which individuals reproduce
process of artificial selection
select population that displays variation
select individuals with desired traits
selectively breed individuals together that display desired traits
grow and test offspring for desired traits
repeat selection process across many generations
issues with artificial selection
inbreeding:
decreases genetic diversity
can lead to expression of harmful recessive alleles
can lead to inbreeding depression→ loss of ability to survive and grow
reduces fitness and stability
outbreeding
breeding unrelated individuals
increases genetic diversity
reduces expression of harmful recessive alleles
can lead to hybrid vigour→ increased ability to survive and grow well
increases fitness and adaptability