lec 1.4 - mutations
1/36
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
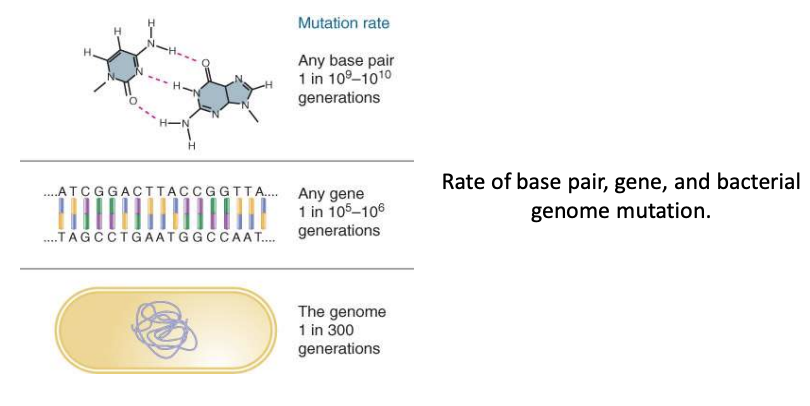
mutations are
periment charges in DNA seq
mutations may occur
spontaneously or may be induced by mutagens
chemically or naturally induced
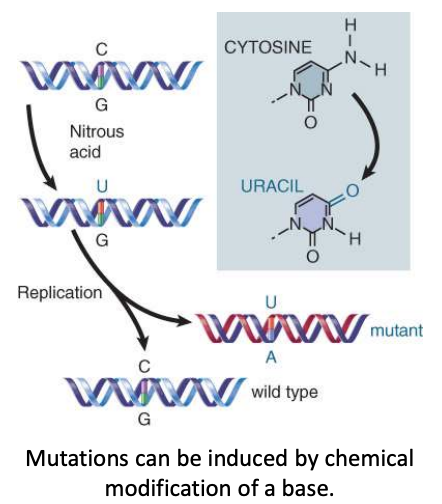
point mutation
change in single base pair
can be caused by the chemical conversion of one base into another or by errors that occur during replication
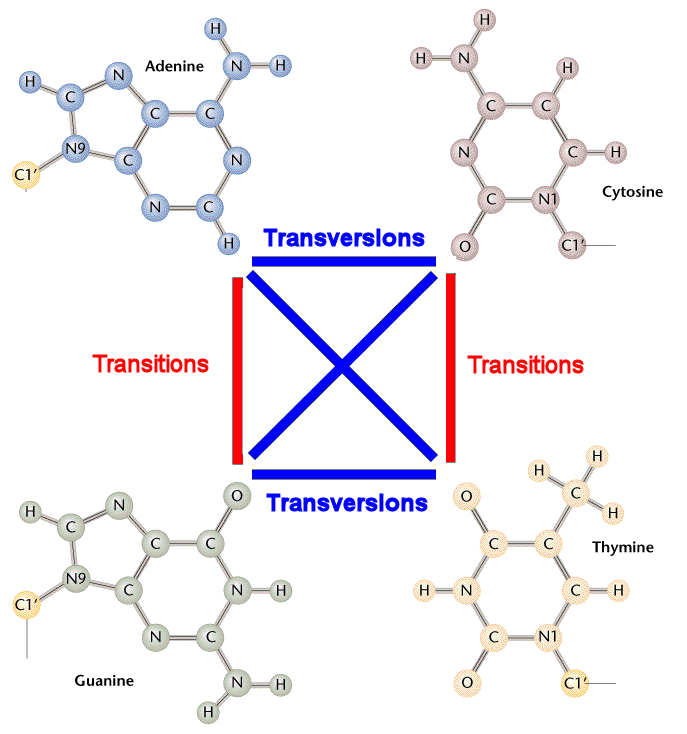
transition and transversion mutations
transition: replaces purine with purine or pyrimidine with pyrimidine
i.e. G-C base pair w A-T base pair or vice versa
transversion: replaces a purine with a pyrimidine
changing A-T to T-A
worse than transition
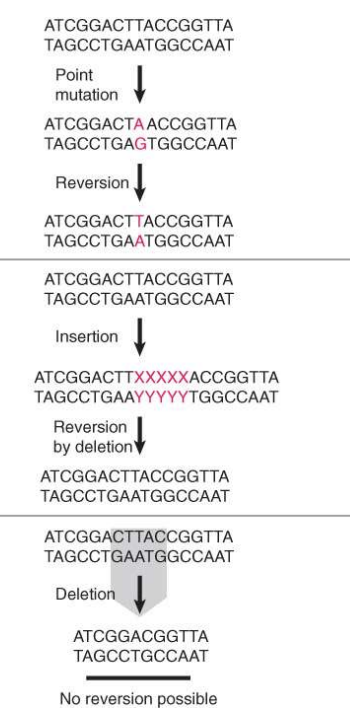
insertions and deletions
addition or deletion of bases and can result from the movement of transposable elements or certain chemicals
insertions can revert by deletion of the inserted material, but deletions cannot revert
forward mutations
alter gene function
back mutations
aka revertants, reverse the effects of forward mutations
back to original sequence
hard/rare
true reversion
mutation that restores the original sequence of the DNA
very rare
second-site reversion
second mutation (at a second site) suppressing the effect of a first mutation within the same gene
suppression
when a mutation in a second gene bypasses the effect of mutation in the first gene
usually when 2 diff proteins interact w/ each other
loss and gain of function mutations
recessive mutations are due to loss of function by the polypeptide product
dominant mutations result from a gain of function
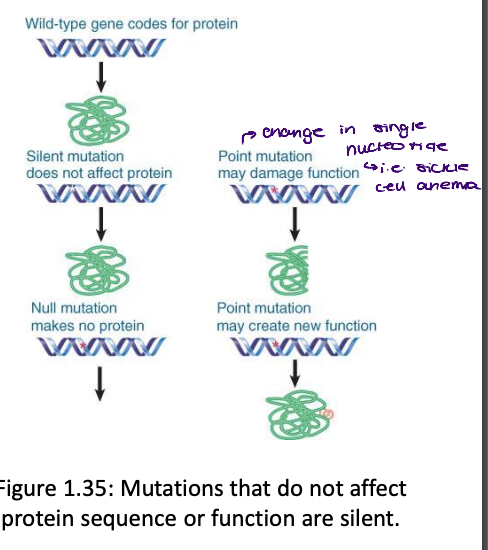
what is required to test whether a gene is essential to life
testing whether a gene is essential requires a null mutation (one that completely eliminates its function).
need to completely knock it out/no functional protein to see if it is needed for a viable organism
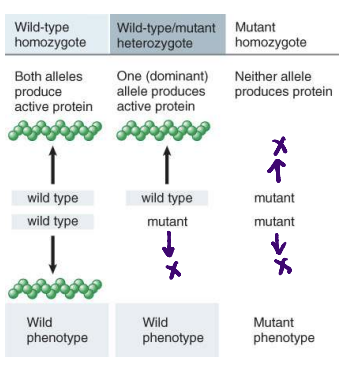
mutations are usually ______ to the wild-type allele
recessive to the wild-type allele
mutations damage gene function
genes encode proteins; dominance is explained by the properties of mutant proteins
complementation test
test that determines whether two mutations are alleles of the same gene
cross two different recessive mutations that have the same phenotype
determine whether the wild-type phenotype can be produced
if the mutations are in the same gene then there will be no complementation (mutation shows). if there is complementation=WT phenotype is expressed that means the alleles are on different genes
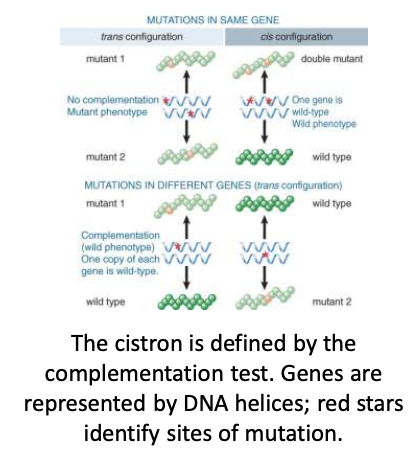
cistron
alternative term for gene
genetic unit defined by complementation test, equivalent to a gene
existence of multiple alleles allows for
the possibility of heterozygotes that represent any pairwise combination of alleles
in other words, the genetic diversity in a population allows for various combinations of alleles to exist in individuals
i.e. in drosophila, many recessive mutations that read to diff eye colours all the way to no colour

polymorphic distribution
when a locus has a distribution of alleles where no individual allele can be considered to be the sole wild type
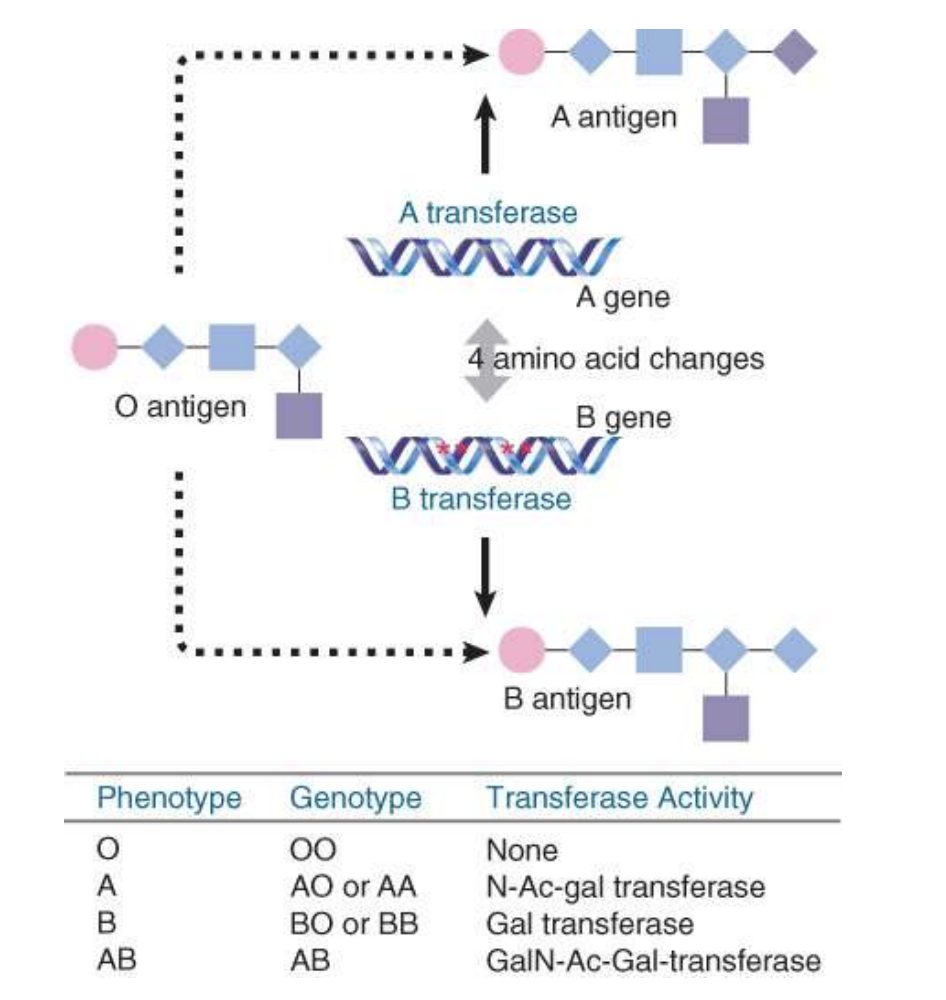
codominance/both alleles ae expressed
i.e. ABO human blood group locus encodes a galactosyltransferase whose specificity determines the blood group
inheriting two A alleles (AA) or one A one O (AO): blood type A, expressing the A antigen on their red blood cells
inheriting two B alleles (BB) or one B one O (BO): blood type B, expressing the B antigen
inheriting A allele and one B allele (AB): blood type AB, expressing both A and B antigens
if they inheriting two O alleles (OO): blood type O, which does not express A or B antigens
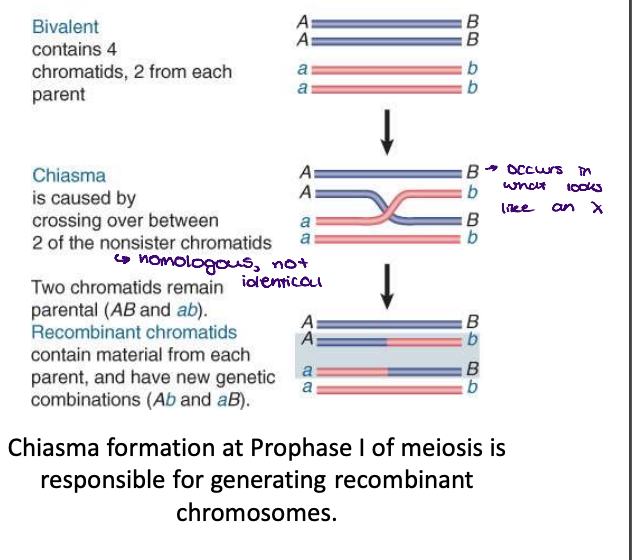
recombination
when in meiosis does this occur?
result of crossing over that occurs at a chiasma (pl. chiasmata) during meiosis and involves two of the four chromatids (two non-sister chromatids)
at prophase 1
no difference if sister chromatids cross (because they are identical), must be non-sister
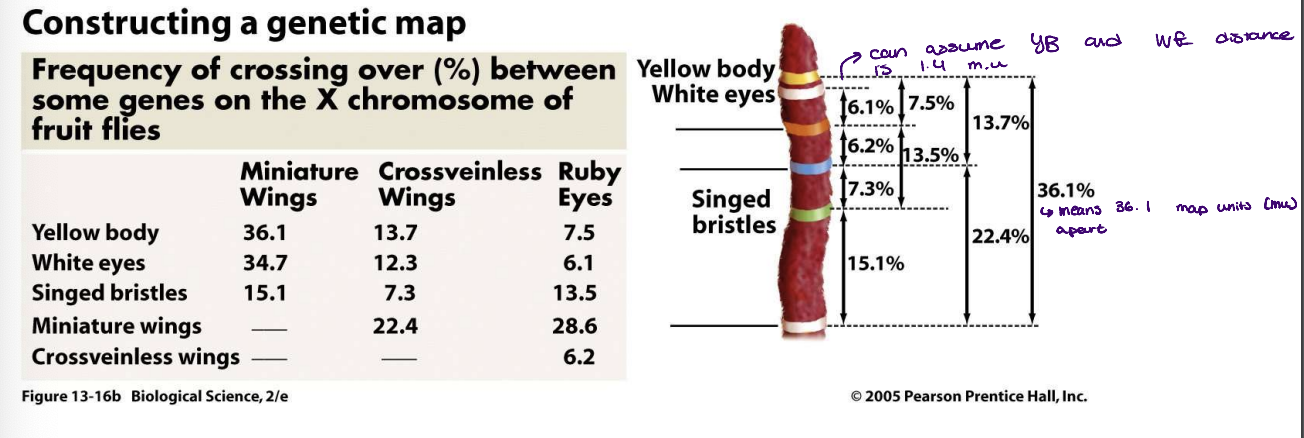
frequency of recombination between two genes is proportional to
their physical distance - used to map the distance between genes
recombination between genes that are very closely linked is rare because they are always inherited as a single unit therefore no recombination
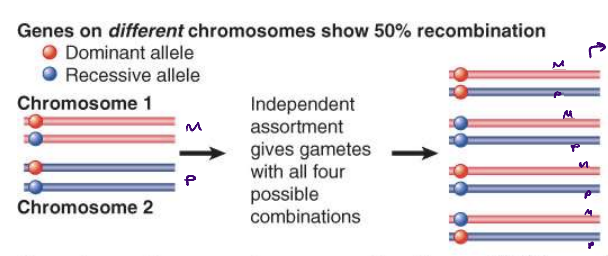
recombination in genes on diff chromosomes
genes on different chromosomes show 50% recombination
recombination due to independent assortment
genes on different chromosomes segregate independently so that all possible combinations of alleles are produced in equal proportions (25% each combination)
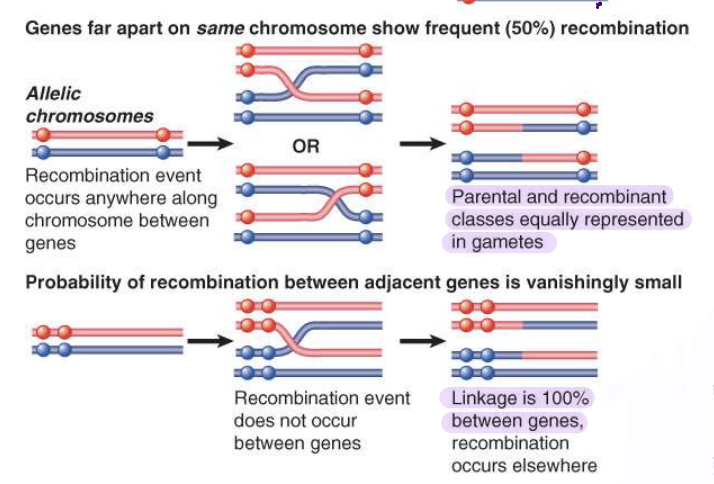
recombination in genes on same chromosome
genes may or may not segregate depending on their distance from one another
50% recombination if genes are far apart
linkage is 100% between genes if they are close on the chromosomes
recomb. elsewhere between other genes, just not with respect to genes under observation
in terms of distance between genes, what is more accurate in determining the recombination frequency and why?
smaller distances are more accurate because the larger ones have multiple sites of crossing over
codons
start codon
triplet nucleotides that the genetic code is read in
triplets are nonoverlapping and are read from a fixed starting point
start codon: AUG
frameshift mutations
mutations that insert or delete 1 or 2 individual bases causing a shift in the triplet sets after the site of mutation
combinations of mutations that together insert or delete three bases (or multiples of three) insert or delete amino acids, but do not change the reading of the triplets beyond the last site of mutation
protein can still fold and be functional (just might do so differently now)

open reading frame (ORF)
usually only one of the three possible reading frames is translated (the one starting at base 1) and the other two are closed by frequent termination signals (usually the ones starting at base 2 and 3)
ORF is a DNA sequence consisting of triplets that can be translated into amino acids, starting w/ initiation codon (AUG) ending w/ term. codon (UAA, UAG, UGA)
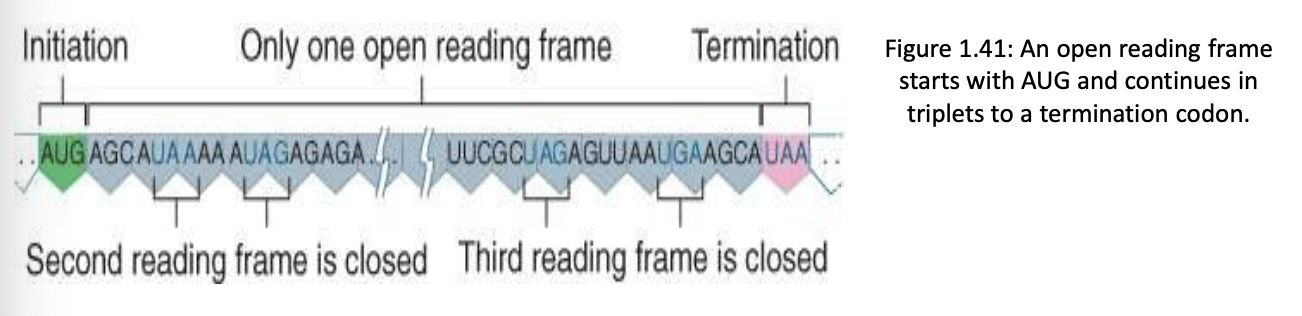
closed (blocked) reading frame and unidentified reading frame (URF)
closed (blocked) reading frame – reading frame that cannot be translated into polypeptide because of the occurrence of termination codons
shorter protein
unidentified reading frame (URF) – open reading frame with as yet undetermined function
looks like an ORF but we are not sure what it codes yet
gene expression
the process by which the information in a sequence of DNA in a gene is used to produce an RNA or polypeptide, involving transcription and (for polypeptides) translation
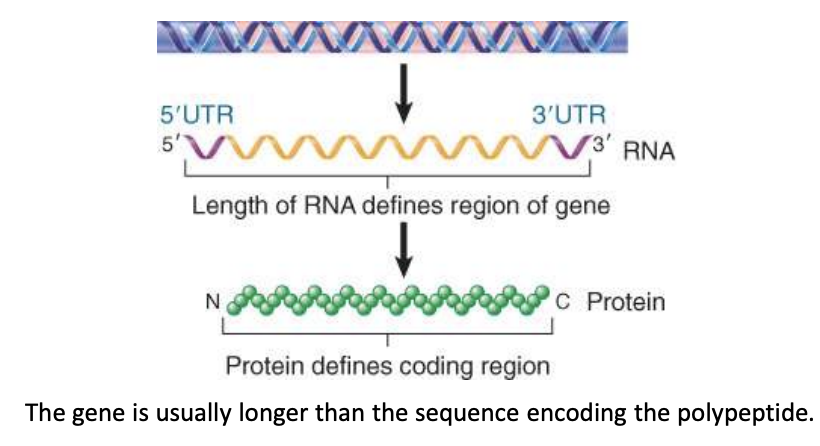
mRNA regions
each mRNA consists of a:
untranslated 5′ region (5′ UTR or leader)
has no codons
coding region
codes for protein
untranslated 3′ UTR or trailer
has no codons
introns
parts of the gene with codons that are not represented in the polypeptide product as they do not code for anything
introns are removed from the pre-mRNA transcript by splicing to give an mRNA that is collinear w/ the polypeptide product
not present in mature mRNA
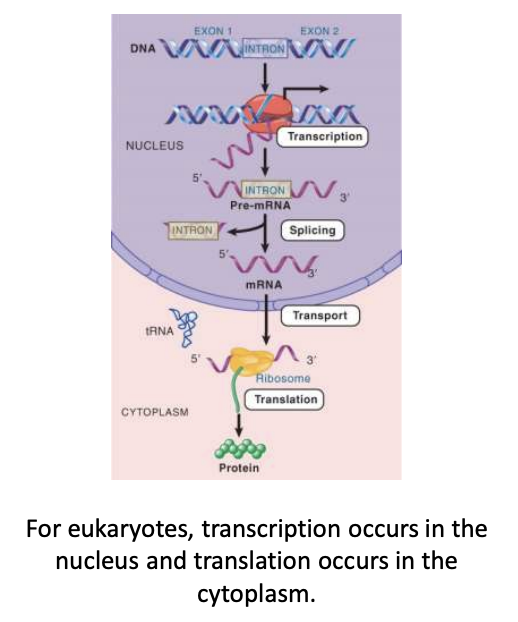
RNA processing
modifications to RNA transcripts of genes.
This may include alterations to the 3′ and 5′ ends and the removal of introns
pre-mRNA
the nuclear transcript that is processed by modification and splicing to give an mRNA (nuclear RNA)
exon
any segment of an uninterrupted gene that is represented in mature RNA product
ribosome
large assembly of RNA and proteins that synthesizes polypeptides under direction from an mRNA template
combo of rRNA and RNA proteins
ribosomal RNAs (rRNAs)
major component of the ribosome
transfer RNA (tRNA)
intermediate in polypeptide synthesis that interprets the genetic code
each tRNA molecule can be linked to an amino acid
tRNA has an anticodon seq. that is complementary to a triplet codon representing the AA
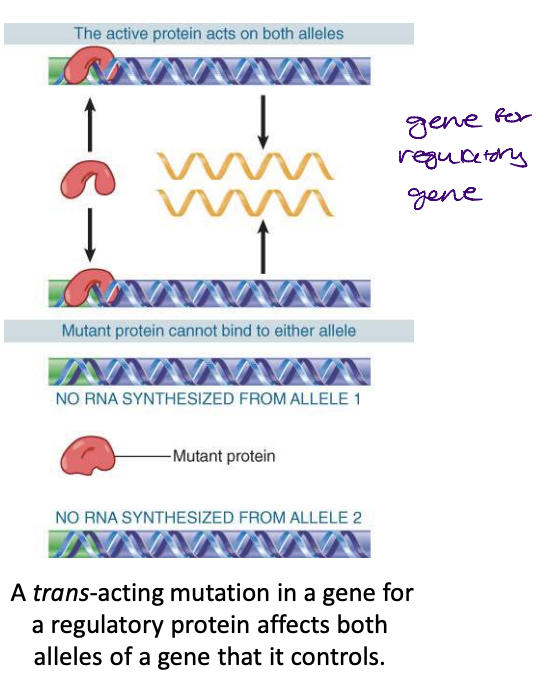
proteins are ____-acting
what are ___-acting mutations
trans-acting : regulatory proteins that bind to DNA
trans-acting mutations identify sequences of DNA that produce proteins that bind to other DNA sequences to cause an effect
trans-acting mutations are expressed as RNA or polypeptide
control sites in DNA provide binding sites for proteins; coding regions are expressed via the synthesis of RNA
sites on DNA are ___-acting
what are ___-acting mutations
cis-acting: DNA sequences in the vicinity of the structural portion of a gene that are required for gene expression.
a promotor
cis-acting mutations identify sequences of DNA that are targets for recognition by trans-acting products
they are not expressed as RNA or polypeptide and affect only the contiguous stretch of DNA
contiguous: multiple DNA sequences next to each other
