2. viral replication
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
virus replication cycle
1. Attachment
2. Penetration
3. Uncoating
4. Transcription “mRNA production”
5. Translation
6. Genome Replication
7. Assembly
8. Release
attachment
binding of the virion to host cells
Glycoprotein spikes attach to receptor on host cell
Must recognize and attach to trigger entry/penetration into the cell
Viral Attachment protein
Protein or glycoprotein that attaches to cellular receptor (ON VIRUS)
target of neutralizing antibodies → can block virus from attaching
Receptor
Cellular target that virion binds
Usually specific protein or carbohydrate
Initiates entry by membrane fusion or receptor-mediated endocytosis
penetration
virus gains access to interior of the cell
Membrane fusion (only enveloped viruses)
Receptor-mediated endocytosis (both types of viruses)
Membrane Fusion
at plasma membrane
ONLY in enveloped viruses
Can also fuse membranes in an endosome
Receptor-mediated endocytosis
virus is engulfed by a vesicle after binding to receptors on cell → brought into cell
vesicle can break down by membrane fusion at the endosomal membrane
OR
by simply breaking down inside of the cell
uncoating
Release of the genome from the capsid
Transcription
synthesis of mRNA
viral genes (DNA or RNA) → mRNA
translation
mRNA → protein
replication
production of progeny copies of the virion nucleic acid (DNA/RNA)
assembly
production of new virion fron the newly synthesized components
release
process when new virions leave the cell
6 Basic Steps of Viral Replication
Attachment
Penetration (entry)
Uncoating
Synthesis.
a. Production of mRNA
b. Translation of mRNA into viral proteins (translation done by the cellular
ribosomes ONLY)
c. Replication of the viral genome
Assembly
Release
what type of cells do viruses replicate in?
it depends on the virus, they replicate in specific host cells
Tropism
Target cells that a virus is able to infect
cell has receptors for attachment and allow replication
Tropism for specific species: humans vs. all mammals (Papillomaviruses are highly species-specific)
Tropism for specific tissues: epithelial cells vs. neurons vs. white blood cells (HSV replicates in neurons and epithelium)
Permissive
a cell that can support virus replication
must have:
Proper surface receptors for the virus (Attachment)
Biosynthetic machinery to support the complete replicative cycle of the virus (Transcriptional factors, Posttranslational processing enzymes, etc.)
Nonpermissive cells
do not allow the replication of a particular strain of virus
may:
Lack a receptor, necessary enzyme pathway or transcriptional activator
Express an antiviral mechanism that inhibits replication
SARS-CoV-2 attachment protein + cellular receptor
Attachment protein: Spike
Cellular Receptor: Angiotensin Converting Enzyme II (ACE2)
One notable benchmark for measuring immunogenicity of a vaccine is the
production of ___________.
neutralizing antibodies (can protect from infection + disease if exposed)
central dogma
DNA → RNA → Protein
some viruses do not adhere to this
translation is done by ribosomes of the cell (NOT VIRAL, VIRUSES do not have ribosomes)
What must happen in every productive viral infection?
(basic)
1. Production of mRNA
2. Translation of viral proteins
3. Replication of viral genomes
What must happen in every productive viral infection?
(more specific)
1. Viral mRNA must be made, regardless of the type of viral genomic nucleic acid
2. Virus must use cellular components and energy to synthesize proteins from mRNA
• Proteins for structure of new viruses, building blocks
• Polymerases, enzymes, evasion proteins, etc.
3. Must replicate its genomic nucleic acid to package in new virions
• All progeny virions must contain a copy of the nucleic acid genome to be infectious to a new cell
Baltimore Classification
Classification system on how viral genomes get to mRNA
Viral genomes must make mRNA that can be translated by host ribosomes
• mRNA is ribosome ready - plus (+) strand
• Complement is the negative (-) strand
• Many viruses, but only 7 known types of viral genomes
what _____ dependent ______ polymerase is always required for RNA replication? why?
RNA, RNA bc host cells cannot convert RNA to RNa
+ sense (coding strand) RNA
genome functions as mRNA, proteins made immediately
positive → protein
- sense (non-coding strand) RNA
must make complementary + sense RNA first
polymerases that can be viral or cellular function
DNA dependent DNA/RNA polymerase
DNA dependent DNA Polymerase
Make DNA from DNA template
Often provided by host cell (Cellular DNA replication)
Some viruses encode their own (Parvo, Pox, Herpes)
DNA-dependent RNA Polymerase
Make RNA from DNA template (transcription)
Host cell: RNA Polymerase II (for mRNAs)
Poxvirus RNA Polymerase (extra-nuclear life cycle)
polymerases that are NEVER cellular function
RNA dependent DNA/RNA polymerase
RNA-dependent RNA Polymerase
Make RNA from RNA template
NOT a cellular function (ever), must be encoded by virus
RNA-dependent DNA Polymerase
Make DNA from RNA template
NOT a cellular function (ever) Must be encoded by virus
Reverse transcriptase (HIV, HBV)
polymerase overview pic
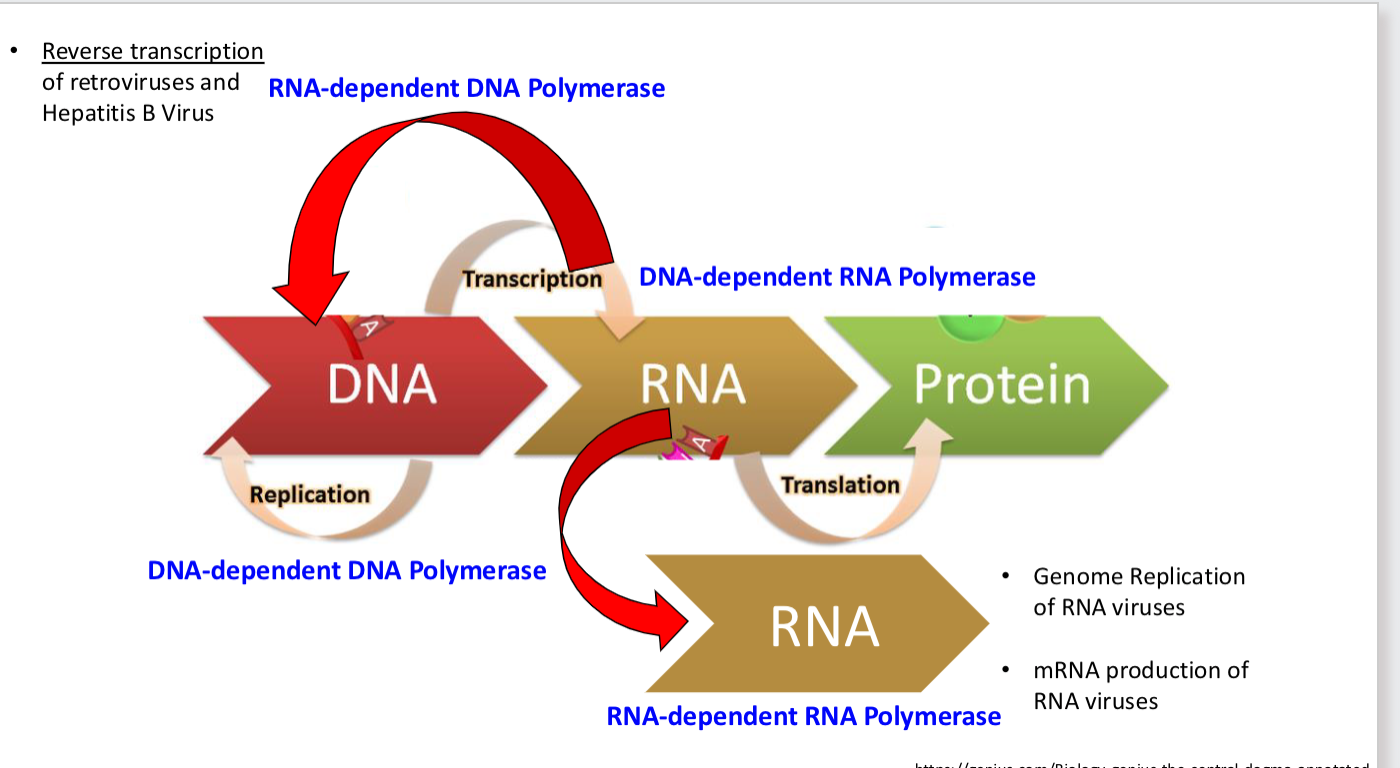
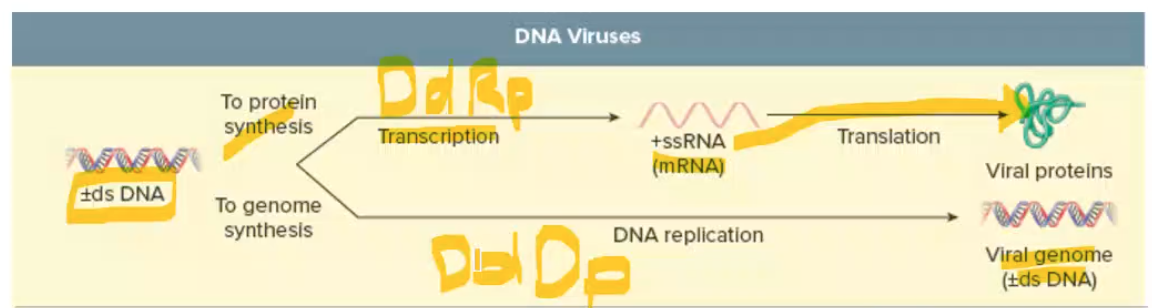
DNA virus replication + transcription polymerases used
Replication of DNA genomes usually occurs in nucleus + uses DNA dep DNA polymerase (DNA genome → DNA genome)
transcription: uses DNA dependent RNA polymerase (DNA genome → protein)
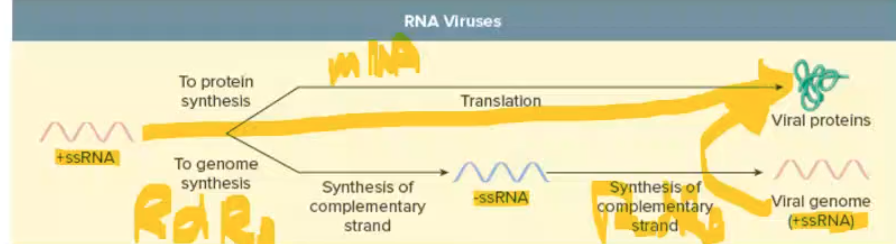
RNA virus replication
Must make and/or bring RNA-dependent RNA polymerase
Mutate faster than DNA viruses bc polymerase has no proofreading (1 million times more than DNA polymerase)
which viruses evolve faster? DNA or RNA?
RNA bc they mutate faster
Plays a role in immune evasion and antiviral resistance
+sssRNA replication + translation
replication: +ssRNA → -ssRNA → +ssRNA uses RNA dep RNA polymerase
translation: +ssRNA = mRNA can become proteins
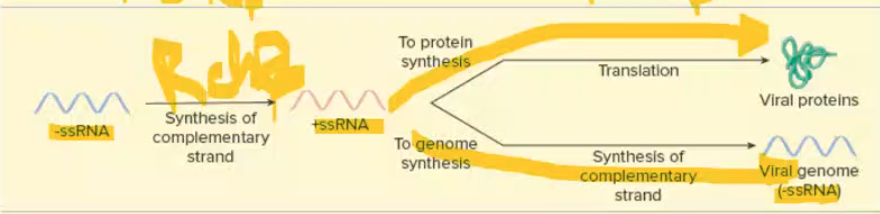
-ssRNA replication + translation
1st step for both: RNA dep RNA poly to become +ssRNA
translation: once +ssRNA = mRNA → proteins
replication: RNA dep RNA poly → RNA (+ssRNA to -ssRNA)
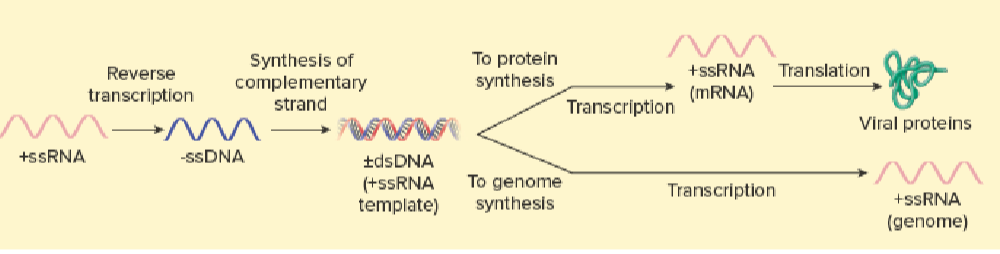
Retrovirus
Retroviridae (HIV)
RNA genome – ssRNA (+) genome
Virion contains reverse-transcriptase (NEVER CELLULAR FUNCTION)
Generates a DNA copy from RNA template (RNA dep DNA polymerase)
DNA integrates into host-cell’s DNA
New RNA genomes are transcribed by host RNA polymerase II (DNA dep RNA polymerase)
RNA → DNA → new RNA
Viral Replication Strategies: Retroviruses
+ssRNA → -ssDNA using RNA dependent DNA polymerase
transcription: DNA dep RNA polymerase (DNA → +ssRNA → translation)
replication: DNA → +ssRNA DNA dep RNA polymerase
Hepatitis B virus
Hepadnaviridae
Circular partially dsDNA genome
DNA is “repaired” by host-cell machinery
Long RNAs are transcribed (pre-genomic RNA,pgRNA)
New partially dsDNA genomes are reverse-transcribed from long RNAs
DNA → (using DNA dep RNA polymerase) RNA → (using RNA dep DNA polymerase) new DNA
SARS-CoV-2: Replication
Family: Coronaviridae
Genome: ssRNA (+), Enveloped
Polymerase: RNA dep RNA polymerase
high likelihood of mutations accumulating
Actual mutation rate is much lower than other RNA viruses like influenza due to exonuclease function of its polymerase
Variants
escape from antibody neutralization, escape from monoclonal antibody therapy, resistance to antivirals
Viral Replication: Release
Budding
Cell lysis
Exocytosis
Budding
capsid and proteins push out of cell membrane taking lipid bilayer with them
ONLY ENVELOPED
Cell lysis
cell bursts open releasing contents
non-enveloped virus, or enveloped (got membrane from organelle inside the cell)
Exocytosis
virus inside the cell has membrane that fuses with the lipid bilayer and gets released
(non-enveloped, enveloped - has two membranes before exocytosis)