cell bio exam 2
1/121
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
122 Terms
what is a chromatin?
Chromatin is the DNA and histone proteins found inside nucleaus that will eventually form into chromosomes.
can either be loose or tightly compacted based on gene activity and cell phase
what is a chromsone
Chromosomes are long, linear strands of DNA tightly packaged with histone proteins.
Highly condensed — ensures DNA is protected and evenly distributed during cell division.
difference betwen chromosome and chromatin
Chromatin = DNA + histone complex that is looser and functional for gene activity.
Chromosome = super-condensed chromatin specialized for DNA packaging and segregation during cell division
Explain how the structure of a chromosome changes as a cell moves from interphase
During interphase, chromatin remains less condensed to allow access to DNA for:
Replication
Repair
Transcription (gene expression)
During mitosis, chromatin fibers fold into tight loops forming highly condensed chromosomes.
This condensation protects DNA from damage and ensures accurate separation of genes during cell divsion
Are all interphase chromosome regions equally condensed
No.
Interphase chromosomes contain two forms of chromatin: heterochromatin and euchromatin, which differ in structure and gene activity
Heterochromatin
highly condensed, found in parts of DNA that do not need to be transcribed (middle and end), genes are not activley expressed and heriable (epigenetic marker)
Euchromatin
most chromosomes, less condensed and have active genes that are transcribed
Heterochromatin vs. Euchromatin
Heterochromatin = silent, structural; Euchromatin = active, gene-rich
Do all cell types have the same heterochromatin regions?
No. All somatic cells have the same DNA sequence, but the regions that are heterochromatic or euchromatic differ by cell type.
This difference reflects cell-specific gene expression patterns (for example, nerve cells silence genes that skin cells express)
Can heterochromatin become euchromatin?
Yes.
Chromosome structure is dynamic.
Chromatin-remodeling complexes use ATP hydrolysis to reposition or slide nucleosomes.
These changes can convert heterochromatin ↔ euchromatin, allowing regulated access to DNA for replication or transcription
What are nucleosomes?
The first level of DNA packaging.
DNA is wrapped around 8 histone proteins
The histone tails have a positive charge (lysine and arginine) so then can bind to negatively charged DNA backbone
what is epigenetics?
chnages in gene function that dont chnage dna seqence, instead changes histone associated with DNA via modifications leading to chnages in level of DNA function and activity
How are nucleosomes regulated?
Histone tails can be post-translationally modified, and these modifications are reversible.
Common chemical groups added to histones:
Acetyl
Methyl
Phosphate
These modifications change the interaction between DNA and histones and attract different non-histone proteins (chromatin-remodeling complexes, HDACs, HATs, transcription factors, etc.)
Histone Acetyltransferase (HAT)
added acetyl group (o-) to tail, causing positive charge to become neutral charge
bond with DNA is loser because there is no longer that positive charge binding to the negative charge
activates gene expression
Histone Deacetylase (HDAC)
removes acetyl group restoring positive charge
strong bond to DNA
closing of DNA=less gene expression
Are histone modifications reversible
yes
What are chromatin remodeling complexes? Why do they require ATP energy?
Protein machines that use ATP hydrolysis to alter packing state of chromatid
This process enables DNA regions to be temporarily exposed or hidden, transition between heterochromatin and euchromatin states
what are the levels of DNA packing
DNA double helix
↓
DNA wraps around histones → nucleosomes (1st level of packing)
↓
Nucleosomes pack together → chromatin fiber (2nd level)
↓
Chromatin folds into loops → visible chromosomes during mitosis (3rd level)
purpose of transcription
The process by which the genetic information stored in DNA is copied into RNA to make a disposable working copy of a gene.
where does transcription occur in eukaryotes
nucleaus
what is the enzyme that carries out transcription
RNA Polymerase II which makes mRNA
function of the three RNA Polymerase
RNA Pol I → rRNA
RNA Pol II → mRNA
RNA Pol III → tRNA and small RNAs
What does “successive amplification” mean
one gene can be transcribed into many RNA molcules and each mRNA can be translated into many proteins
What are the benefits of successive amplification?
Proteins can quickly be synthesized when needed
Transcription and translation efficiency can be different for each gene
Cells can change gene expression at different times (cells don’t want the same protein and amount 24/7)
What do transcription produce?
RNA
coding: mRNA
non coding rRNA, tRna
mRNA function
transferred into protein
non messanger RNA function
do not encode for protein
rRNA: forms ribosome core which translate mRNA into proteins
tRNA: holds amino acid in place on ribosom
What is the difference between coding and templete strand?
Coding strand: Same sequence as RNA (except T → U), not used as template.
Template strand: Strand used by RNA polymerase to synthesize mRNA
In what direction is RNA synthesized?
5′ → 3′ direction, reading the DNA template 3′ → 5′.
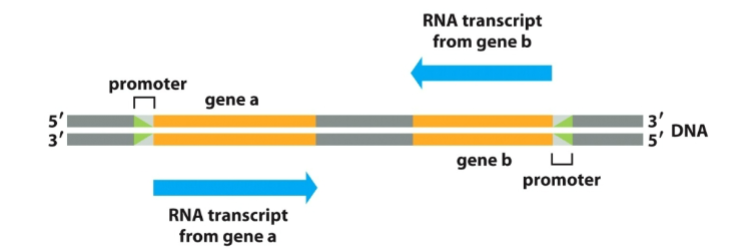
Where does transcription start?
At the promoter, a specific DNA sequence (often containing a TATA box).
What is the role of the promoter?
tells RNA polymerase where to bind and start transcribing, and which strand to use
Can RNA Polymerase initate transcription on its own? Why or why not?
No, needs General Transcription Factors (GTFs) what are proteins that assemble with RNA Pol II at the promoter to help initiate transcription.
Functions of General Transcription Factors?
Help position RNA Pol II at the promoter.
Help separate two strands of DNA
Release RNA Pol II from the promoter to go into elongation mode.
What is the TATA box?
A promoter sequence recognized by TFIID through its TBP (TATA Binding Protein) subunit.
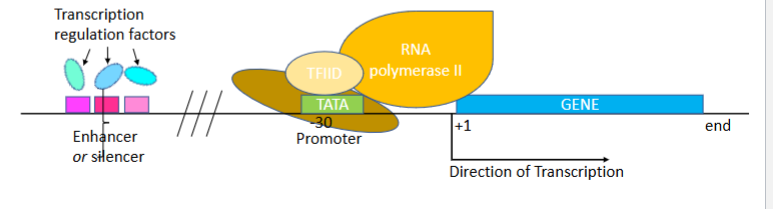
TFIID function
Binds to the TATA box via TBP. Once binded distorrrs DNA double helix and recruits more general transcription factors
TFIIB fuction
Links the promoter, TBP (in TFIID), and RNA Polymerase II to position the polymerase correctly
TFIIH function
Has helicase activity (opens DNA using ATP) and kinase activity (phosphorylates the Pol II tail to start elongation).
What lets RNA polymerase II to being transcription
TFIIH binding which phosphorlates its tail
What happens during elongation of transcription
RNA Pol II moves along the DNA template, synthesizing RNA 5′ → 3′
Give an overview of transcription (long)
Location: Nucleus (eukaryotes).
Start: At the promoter region — often a TATA box.
End: Termination occurs downstream of the AAUAAA signal.
Main enzyme: RNA Polymerase II.
Key factors: General Transcription Factors (GTFs) such as TFIID and TFIIH.
Process:
TFIID binds the TATA box via TBP, bending DNA and forming a landmark.
TFIIB joins → positions RNA Pol II at the promoter.
Other GTFs assemble → form the Pre-Initiation Complex.
TFIIH uses ATP helicase activity to unwind DNA and kinase activity to phosphorylate Pol II.
Phosphorylation allows Pol II to enter elongation → synthesizes RNA 5′ → 3′.
At AAUAAA, mRNA is cleaved and poly-adenylated.
The leftover RNA is degraded by the “torpedo” RNase → RNA Pol II falls off the DNA
What are enhancers?
before promotor, can be every far away
-Distant DNA sites binding activators to increase transcription.
What are silencers?
DNA sites binding repressors to reduce transcription.
How do enhancers/silencers act far away?
DNA looping brings them near the promoter.
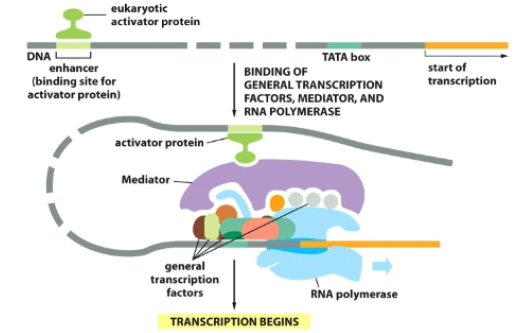
Steps of transcription (story style)
1⃣ TFIID binds TATA → DNA bends.
2⃣ TFIIB positions Pol II.
3⃣ Other GTFs assemble → complex.
4⃣ TFIIH unwinds + phosphorylates.
5⃣ RNA made 5′→3′ → terminated at AAUAAA.
what if TFIID is missing?
the promotor is not recongized so transcription can not occue
what if TFIIH is missing?
DNA can’t unwind so no elongation can occur
What if no HATs?
Chromatin stays closed so the gene is off
transcription one line summary
dna becomes rna via rna polymerase II and general transcription factors
Where does transcription occur in eukaryotes?
nucleus
Where does translation occur in eukaryotes
cytoplasm
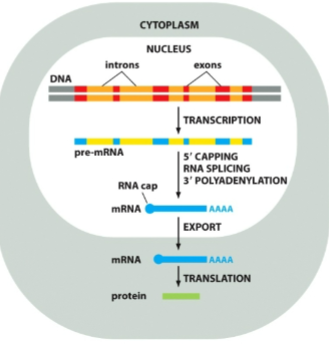
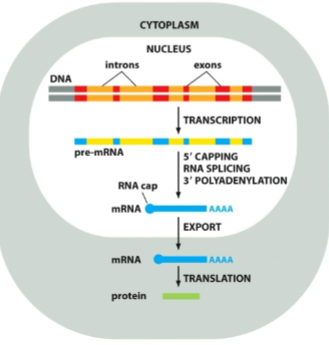
Why do eukaryotic cells modify RNA before being exported to cytoplasm to be translated
protect the molecule from degradation by ribonuclease, which clears old RNA
removed the sequences that don't need to be translated
What must happen to eukaryotic mRNA to become mature and functional
it must be capped, polyadenylated and spliced before being exported to cytoplasm
When do mRNA modifications occur
after transcribtion, before cytoplasm exportation for translation
What are the three main processing steps for mRNA?
5’ cap
poly-a-tail
splicing
What happens during 5′ capping
A guanine nucleotide with a methyl group is added to the 5′ end of the mRNA shortly after transcription begin
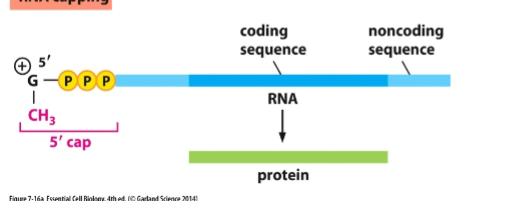
what is the purpose of the 5” cap
protect from degradation
exit signal for mRNA to leave nucleusthe
help ribosome binding
whats happens during addition of 3’ Poly-A tail
adds 100’s of adeneine nucleotides to 3’ end of mRN
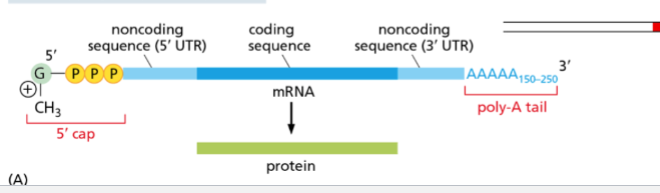
What is the purpose of the poly-A tail
protect 3’ end from degradation
signal to exit nucleus
helps end transcription
What are introns
Non-coding RNA sequences that must be removed before translation
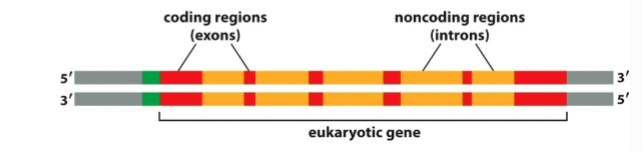
What are exons
coding sequences that stay in mRNA and determine protein sequence
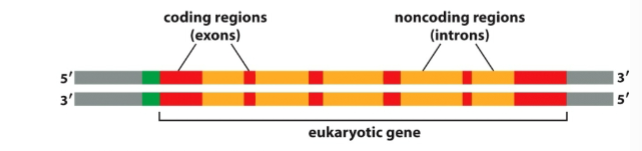
What is splicing and why does it happen
removing introns to create a strand of only extons (coding) that alllows more infomration to be packed into one gene and produce different protein versions
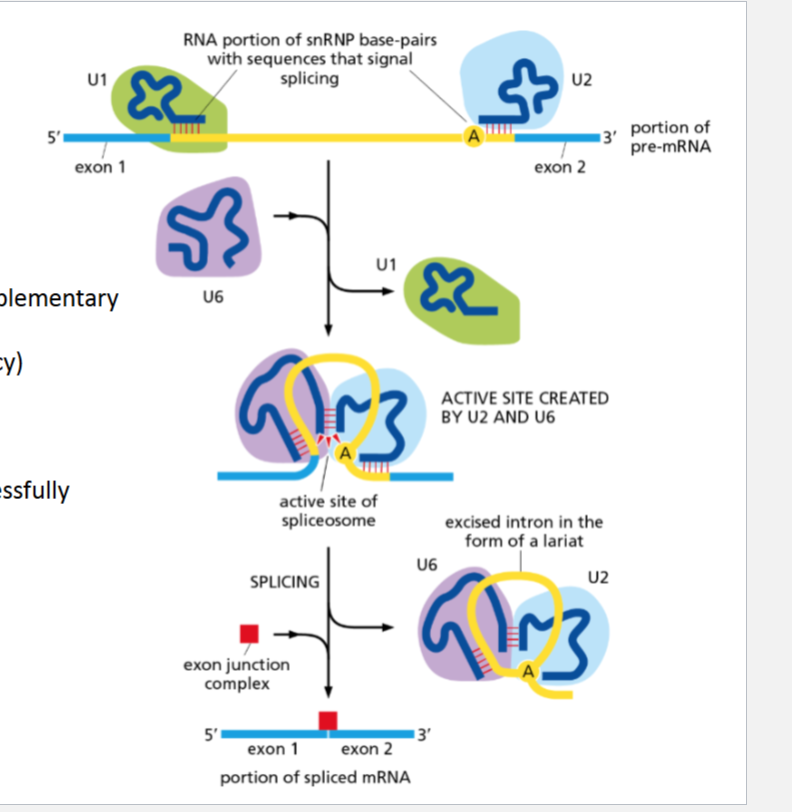
What performs splicing
spliceosome, a large complex of snRNPs (small nuclear ribonucleoproteins) and proteins
How does the spliceosome work?
U1 binds the 5′ splice site.
U2 binds the branch point adenine.
U6 rechecks the 5′ site and forms the active site.
Once the axon joins, the exon junction complex is deposited to signal that splicing is done.
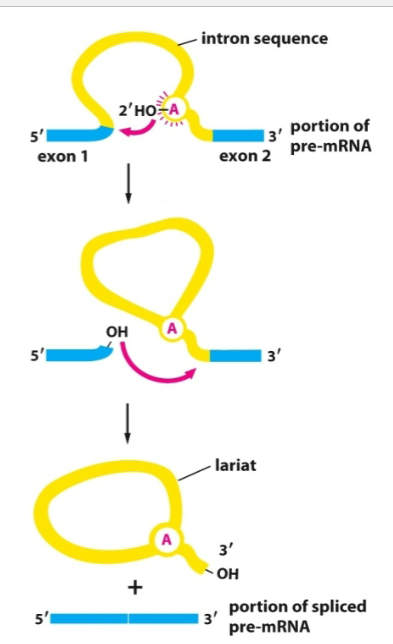
Basic steps of splicing
2’ OH of adenine attacked 5’ splice site
cut of 5’ end of intron is covalently linked to 2’OH end
free 3’ OH of extron reacts w next extron then intron is removed
How does mRNA leave nucleaus?
via nuclear pores
complex gates
recognizes if mRNA is mature, needs…
CAP binding complex: proteins bound to 5’ Cap
Poly-A-binding proteins- proteins bound to the Poly A tail
Exon junction complex: makes sure introns are spliced
Why is RNA less stable than DNA
RNA has a 2′-OH group that makes it more prone to hydrolysis and degradation
Why is splicing advantageous
It allows alternative splicing, where one gene can make multiple mRNAs and proteins depending on cell type or conditions
Quick summary of mRNA processing order
1⃣ 5′ capping → protection & export signal
2⃣ Splicing → remove introns, join exons
3⃣ Poly-A tail → protection, export, translation aid
What is the goal of translation?
convert genetic info in the form of mRNA into protein
Where does translation occur in the cell?
In the cytosol on ribosomes
Can an mRNA be used more than once?
Yes — the same mRNA can be translated repeatedly to make multiple copies of a protein
Can multiple ribosomes translate one mRNA at the same time?
Yes — this forms a polyribosome (polysome), allowing faster, amplified protein synthesis
What are the three main stages of translation
Initiation, Elongation, Termination
What is the goal of translation initiation?
Bind mRNA, locate the start codon (AUG), assemble ribosomal subunits, and begin the first peptide bond
What factors help bind mRNA during initiation?
eIF4E binds the 5′ cap, PABPs bind the poly-A tail, and both connect via eIF4G to circularize mRNA
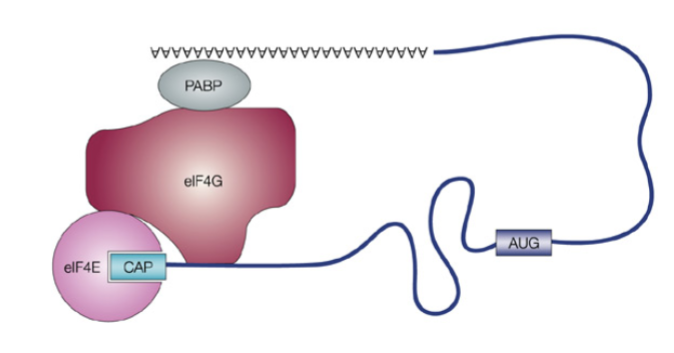
What tRNA starts translation
The initiator tRNA charged with methionine (Met) binds the start codon AUG in the P site
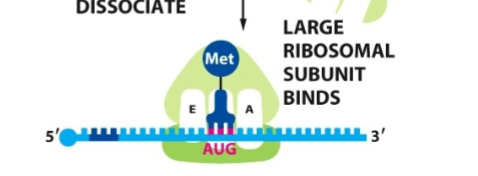
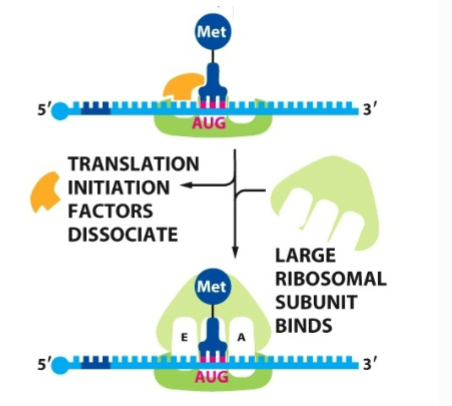
What happens after the start codon is found by small ribosome subunit
initation factors leave and large subunit comes in
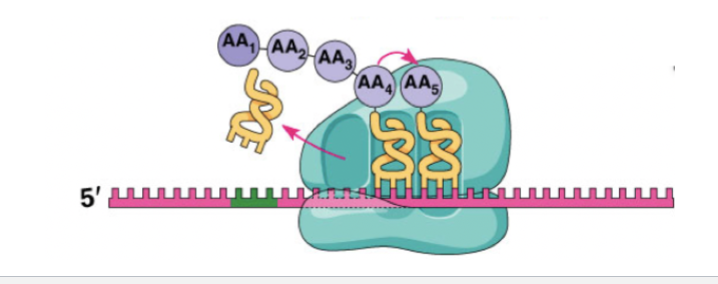
elongation goal
The ribosome moves 5′→3′ along mRNA, adding amino acids to the growing chain one codon at a time
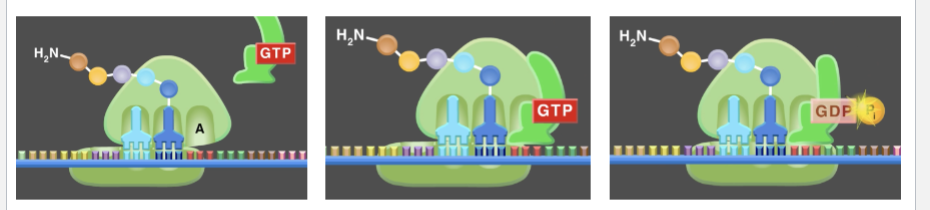
What does Elongation Factor-Tu do (EF-TU)
Brings charged tRNA to A-site (uses GTP).
what does aminoacyl-tRNA do
links each tRNA to its correct amino acid
What is a peptidyl-tRNA
tRNA in P site that carries growing poly peptide chain
What does EF-G do
moves small subunit foward via gtp hydroysis
What are the ribosome’s three tRNA binding sites
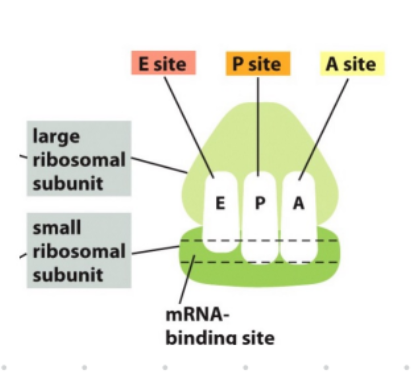
A site: “charged” tRNA enters w amino acid
P site: holds peptide chain and where first tRNA meets start codon
E site: exit site for empty tRNA
Steps of elonglation
correct tRNA added to the A site
peptide bond forms between an amino acid at the P site and the A site (peptidyl transferase catalyzes)
large subunit moves and everything, shifts, and empties the A site for the new codon
empty tRNA ejected from the site
keeps adding more amino acids
what is the goal of termination
Release the complete polypeptide chain
recycle the machinary
What signals termination
A stop codon (UAA, UAG, UGA) with no matching tRNA
terminination steps
ribosome arrives at stop codon
realease factor binds to A site
conformation chnage in large subunit
water is added to tRNA instead of amaino acid
polypeptide chain and other factors realsed
what is a codon
A three-nucleotide sequence in mRNA that specifies one amino acid
what is the start codon
AUG, which codes for methionine
What is an anticodon
The complementary three-nucleotide sequence on tRNA that base-pairs with the mRNA codon
basics of ribosome structure
has a small subunit (decodes mRNA)
large subunit (forms peptide bonds)
mRNA binding site
A, E, P site
what is tRNA
a small RNA that matches a codon with corresponding amino acid
what does amino acyl synthase do?
creates bond with tRNA and amino acid
What is a polyribosome (polysome)
Multiple ribosomes translating the same mRNA simultaneously
summary of translation steps
Initiation: Assemble ribosome at start codon
Elongation: EF-Tu delivers charged tRNAs; EF-G moves ribosome; peptidyl transferase links amino acids
Termination: Stop codon → release factor → peptide release & ribosome recycling
what are stem cells
undifferentiated cells that can devloped into different specialized cells
What is the role of stem cells in development
they divide and become differnt by turning specifc genes on or off so then speicalized tissues can be devlop
What are transcriptional regulators?
They activate or repress proteins that control transcription by binding specific nucleotide sequences in DNA (not the backbone) at enhancer or silencer regions near promoter
why are transcriptional regulators called “master regulators”?
Because a small number can control expression of many genes. Combinations of just a few can produce many different cell types
What is meant by “combinatorial control”?
many regulators work together to determine if a gene is expressed