WJEC AS Biology Unit 1.5 - Nucleic acids and their function
1/109
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
110 Terms
Nucleotides
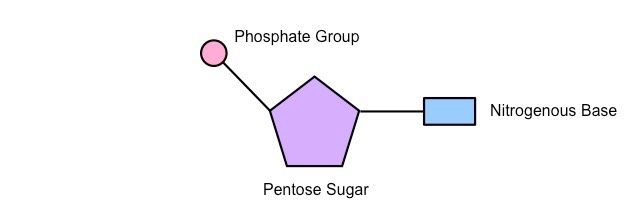
Monomer of nucleic acid comprising a Penrose sugar, a nitrogenous base and a phosphate group
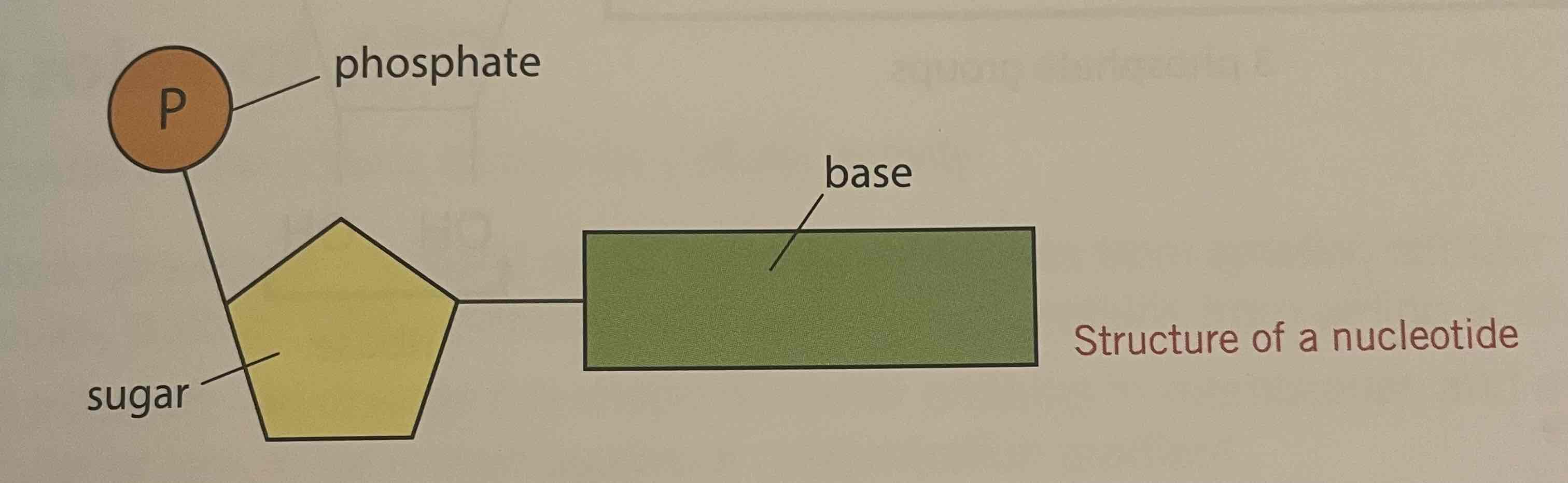
Structure of nucleotides (description)
Three components, combined by condensation reactions;
A phosphate group; which has the same structure in all nucleotides
A pentose sugar; in RNA = ribose and in DNA = deoxyribose
An organic, nitrogenous base
Structure of nucleotides (diagram)
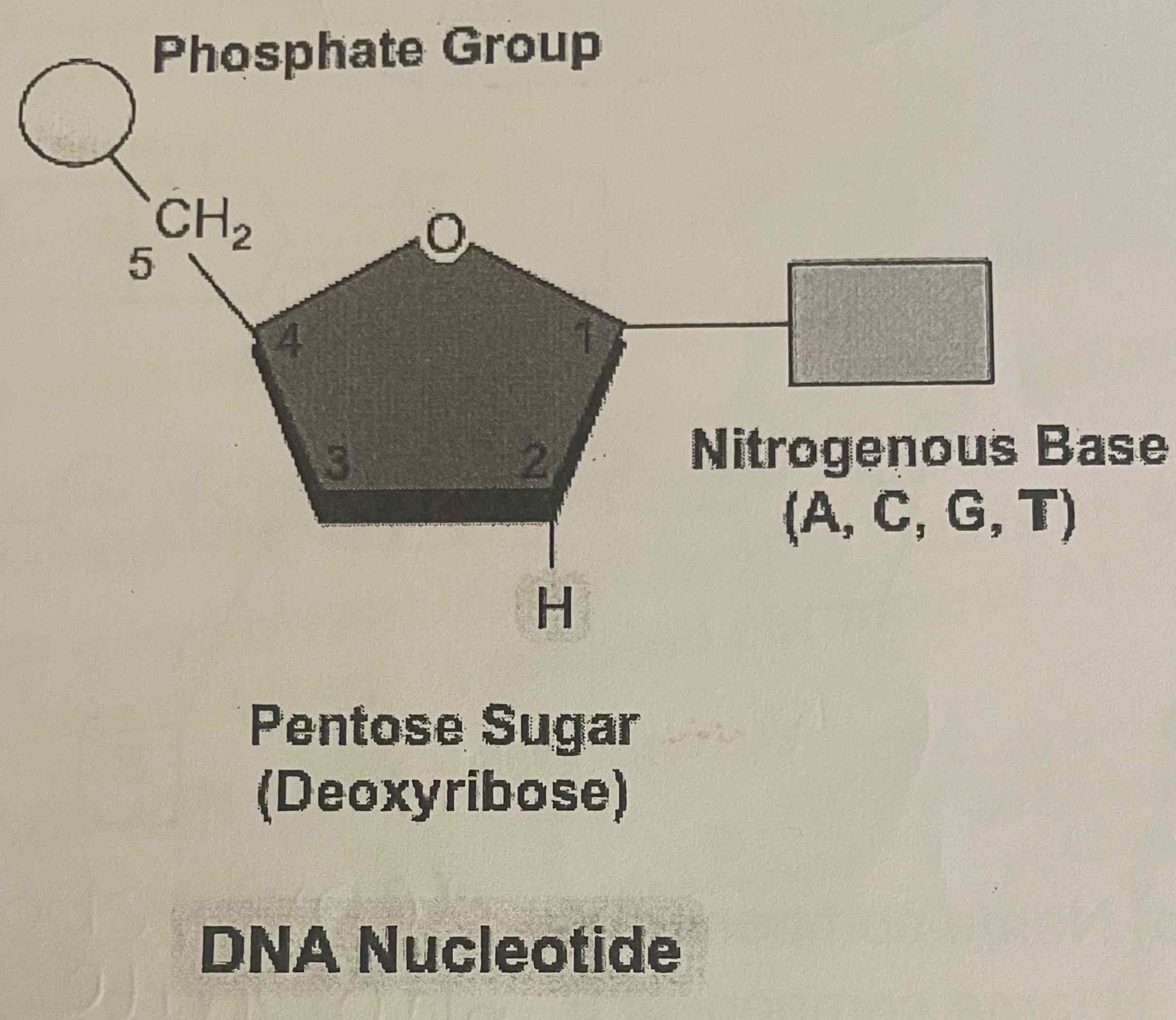
DNA nucleotides
Phosphate group
Deoxyribose pentose sugar
Nitrogenous base (A, C, G, T)
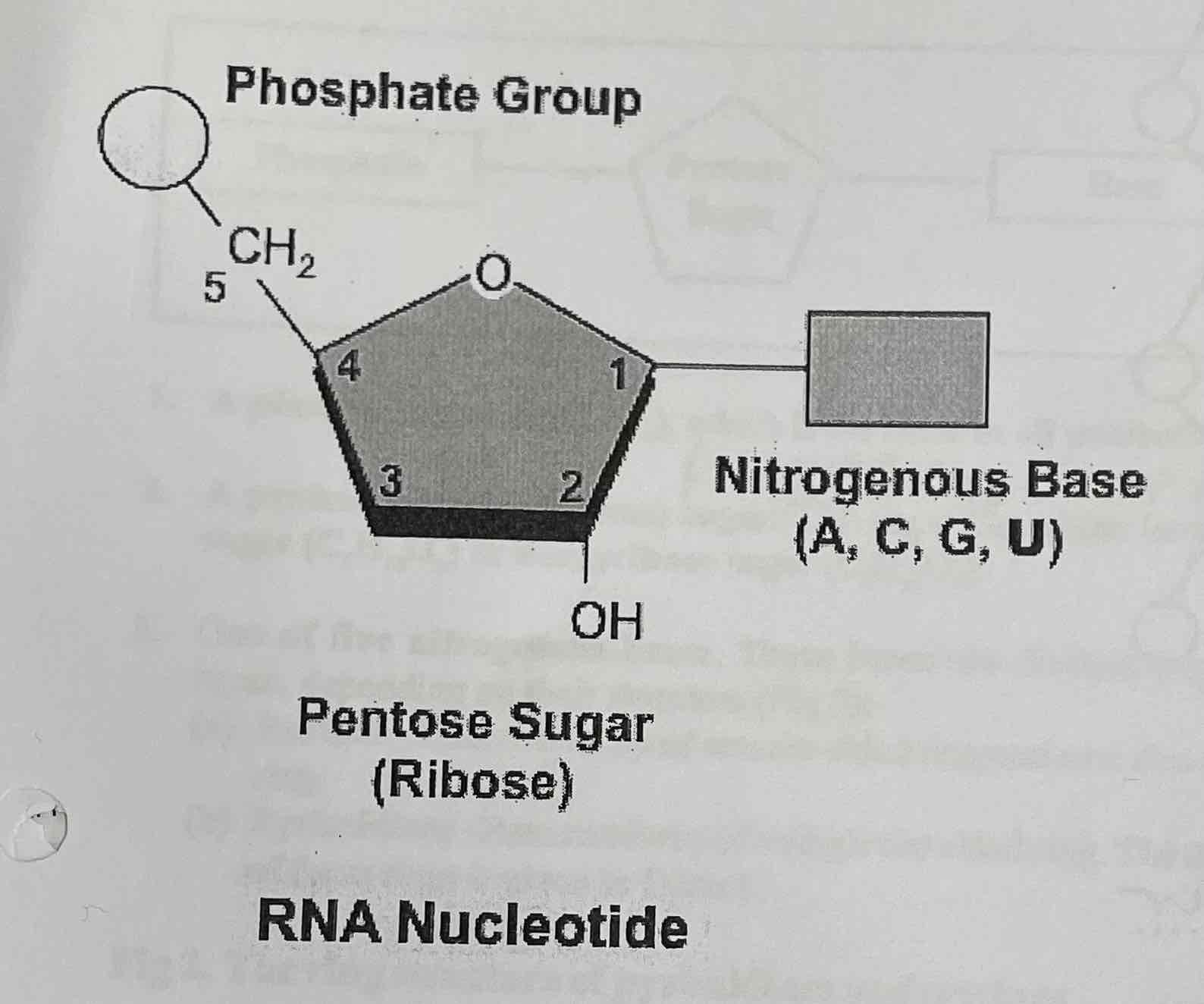
RNA nucleotides
Phosphate group
Ribose pentose sugar
Nitrogenous base (A, C, G, U)
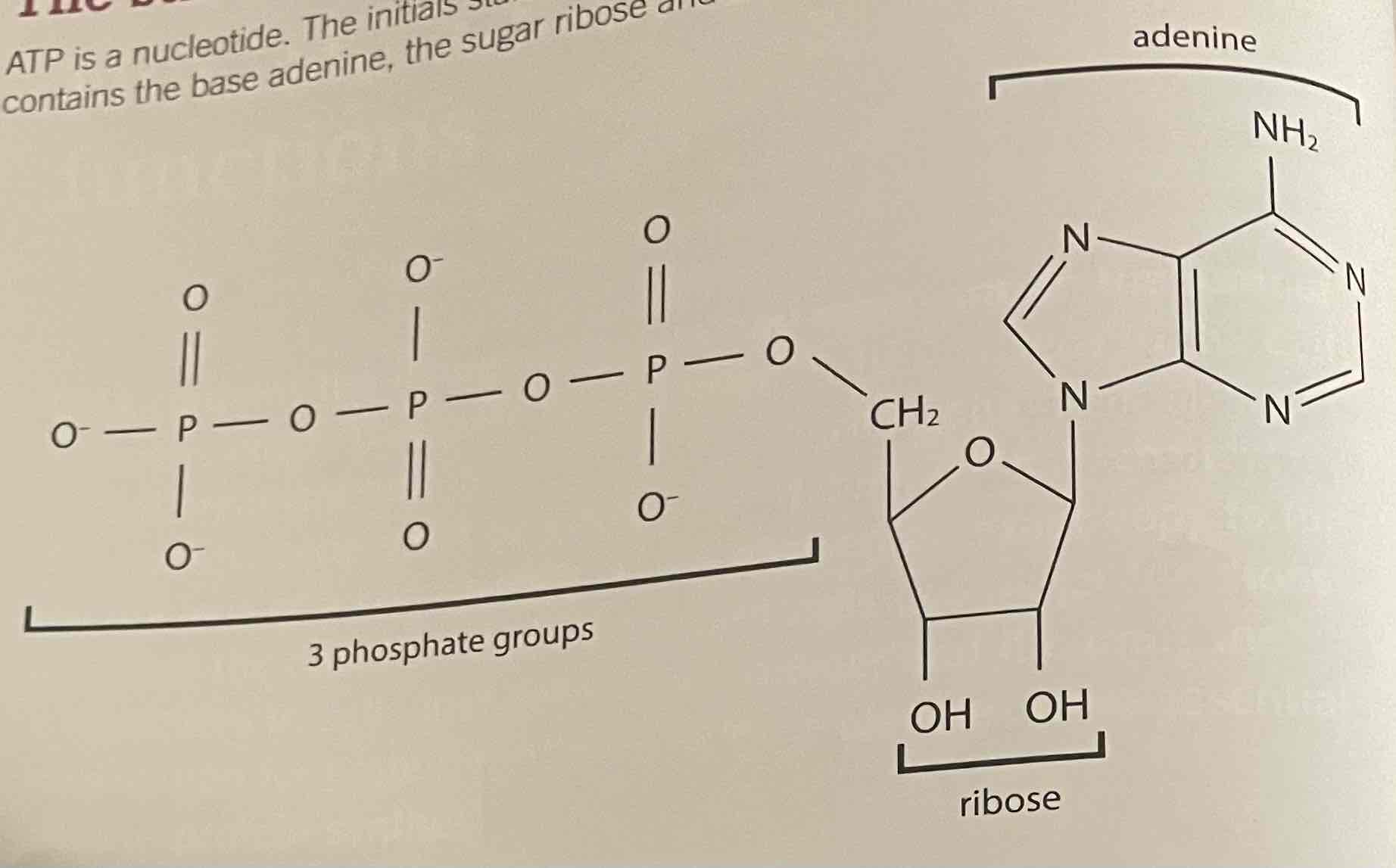
ATP nucleotides
3 Phosphate groups
Ribose pentose sugar
Adenine nitrogenous base
ATP as an energy carrier
Known as the universal energy currency as it is a common energy source/energy carrying molecule used in all cells in all living organism
ATP synthesis
Formed when ADP (adenosine diphosphate) is combined with a third phosphate group (Pi; inorganic phosphate)
Endergonic reaction as it requires energy to be taken in
The energy is then trapped in bonds between the phosphate groups
ADP is phosphorylated to ATP, which means it gains an inorganic phosphate group
ADP + Pi + Energy —> ATP + Water
Condensation reaction
Catalysed by enzyme
Hydrolysis of ATP
energy is released
ATP is hydrolysed to ADP and Pi
Exertions reaction as it releases energy
30.6 kilojoules of energy is released per mole of ATP
Energy required to combine ADP and inorganic phosphate to form ATP and water
comes from exerting reactions, e.g. cell respiration
ATP as the “universal energy currency”
common energy source used in all biochemical reactiosn in all living organisms
Phosphorylation
The addition of a phosphate group
Advantages of ATP as a supplier of energy/energy carrier molecule
Hydrolysis of ATP to ADP and Pi = single reaction that releases energy immediately. Breakdown of glucose requires many intermediates and takes longer for energy to be released
One enzyme needed to release energy from ATP (ATP Synthase). Many needed to release energy from glucose glucose
More manageable amount of energy released
Easily regenerated from ADP and Pi
Provides common source of energy for all chemical reactions —> increasing efficiency + control in the cell
Small molecule —> can easily diffuse throughout the cytoplasm
Uses of ATP
Metabolic processes
Active transport
Movement
Nerve transmission
Secretion
Cell division
DNA
DNA is made from one strand of nucleotides linked by hydrogen bonds between the bases to another strand that runs antiparallel to the first.
composed of 2 polynucleotide strands wound around each other —> double helix structure
Pentose sugar in nucleotides = deoxyribose
Four organic bases = 2 purines; adenine and guanine + 2 pyrimidines; cytosine and thymine
Sugar phosphate back bone formed by deoxyribose sugar and phosphate groups
Complementary base pairing; A+T and C+G
Hydrogen bonds join the bases, forming complementary pairs + maintaining the shape of the double helix
Adenine + Thymine joined by 2 Hydrogen bonds
Cytosine + Guanine joined by 3 hydrogen bonds
Antiparallel strands in DNA
The 2 polynucleotide strands antiparallel
The nucelotides in 1 strand are arranged in the opposite direction from those in the complementary strend
parallel but facing in opposite directions
Run in opposite directions; one from the 5’ prime end to the 3’ prime end, the other from the 3’ prime end to the 5’ prime end
Pyrimidine base
Bases made up of a single six-sided ring
Class or nitrogenous bases including Cytosine and Thymine in DNA and Cytosine and Uracil in RNA
Purine bases
Bases made up of one six-sided ring and one five-sided ring
Class of nitrogenous bases including adenine and guanine in both DNA and RNA
Bond between nucleotides in the backbone
Phosphodiester bonds
Bonds between bases in DNA
Hydrogen bonds;
2 between A and T
3 between C and G
RNA
single stranded polynucleotide
Contains the pentose sugar ribose
Contains the nitrogenous bases Adenine + Guanine (purines) and Cytosine and Uracil (pyrimidines)
Three types;
mRNA; made as a complementary copy of the DNA genetic code in the nucleus during transcription. The molecule length is related to the length of the gene transcribed. It attaches to a ribosome in the cytoplasm. long, single stranded molecule. Synthesised in the nucleus + carries the generic code from DNA to ribosomes in cytoplasm
rRNA; found in the cytoplasm + comprises large, complex molecules. forms ribosomes
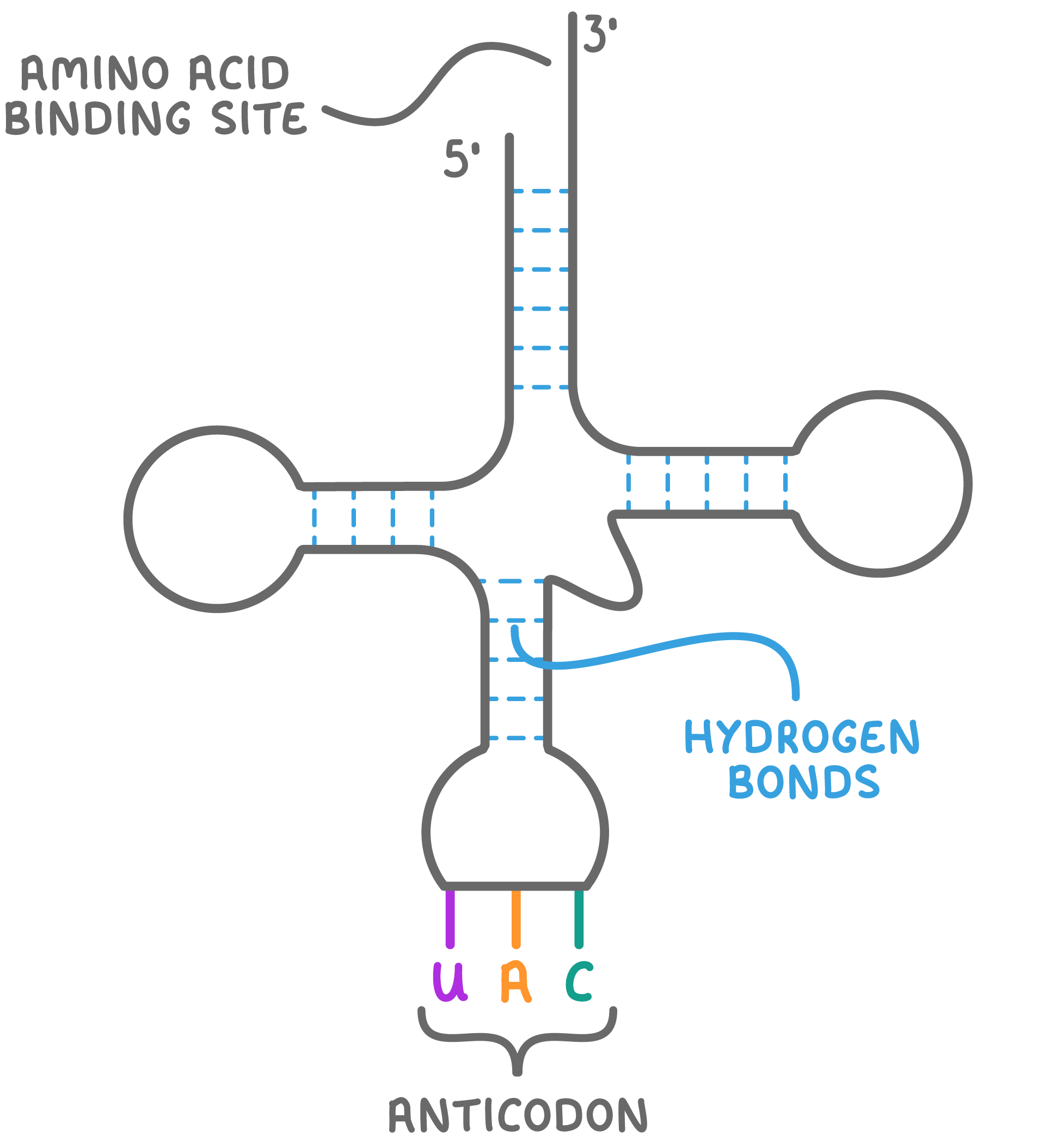
tRNA; carries an amino acid at the 3’ end and an anticodon arm to attach to the mRNA. small single stranded molecule. Folds so that, in places, there are base sequences —> complementary pairs
tRNA structure
The 3’ end of the molecule has the base sequence cytosine-cytosine-adenine where the specific amino acid the molecule carries is attached
Also carries a sequence of 3 bases = anticodon
molecules of tRNA transport specific amino acids to the ribosomes in protein synthesis
Properties, structure and formation to function of ATP
ATP releases energy in one hydrolysis reaction controlled by one enzyme.
ATP releases energy in small, usable amounts.
ATP travels easily to where it may be used for secretion, muscle contraction, nerve transmission or active transport.
How DNA is suited to its functions
very stable molcule and ita information content passes essentially unchanged from generation to generation
very large molcule and carries a large amount of genetic information
the 2 strands are able to seperate, as they are held together by hydrogen bonds
as the base pairs are on the inside of the double helix, within the deoxyribose-phosphate backbones, the genetic information is protected
Major functions of DNA
Replication
DNA comprises 2 complementary strands, the base sequence of 1 strand determining the base sequence of the other. if 2 strands of a double helix are separated, 2 identical double helices can be formed, as each parent strand
Protein synthesis
sequence of bases represents the information carried in DNA and determines the sequence of amino acids in proteins
Comparison of DNA and RNA
Semi-conservative replication of DNA
Each of the strands of DNA acts as a template to make a new strand. New DNA molcules are made of an original strand linked to a new strand
Meselson and Stahl carried out an experiment which gave evidence to the theory of semi-conservative replication of DNA.
Conservative replication
bpth parent strands stay together and the parental double helix remains intact/is conserved and a whole new double helix is made
direct copying of the nucleotide sequence onto a new double stranded molecule which would give one light and one heavy molecule in generation 1.
Dispersive replication
sections of old and new DNA are found in sections all strands so all copies would be intermediate density
where half the nucleotides are placed randomly in the DNA being replicated to make new molecules which would give successively lighter molecules and therefore a band between hybrid and light in generation 2.
Meselsohn and Stahl Experiment
Gives evidence of the semi-conservative theory of DNA replication and disproves other theories; dispersive and conservative
Grow bacteria with a heavy isotope of nitrogen. Centrifuge a sample – a heavy band is seen.
Remove bacteria with heavy DNA and place into a medium with light nitrogen and allow bacteria to divide. They will synthesise DNA with the nitrogen isotope available meaning their DNA will contain 1 new (14N) and 1 old (15N) strand, making it intermediate in density when centrifuged.
Allow 1 more generation to grow and the hybrid strands will now be copied in a semi conservative way creating 50% hybrid and 50% light DNA
Enzymes in DNA replication
DNA Polymerase
DNA Helicase
Process of DNA replication
Replication starts at a specific sequence on the DNA molecule; the replication origin
DNA helicase unwinds and unzips DNA, breaking the hydrogen bonds that join the complementary base pairs, forming 2 separate strands
Bases are now exposed and unpaired
Free nucleotides in the nucleoplasm are bound to their complementary bases on the unzipped strand.
DNA polymerase joins the nucleotides together by condensation reactions between sugar and phosphate groups of adjacent nucleotides, forming phosphodiester bonds in the new strands of DNA being formed
A winding enzyme winds the new strands up to form double helices
2 new DNA molecules are formed from 1 old molecule and 1 new molecule. The 2 new molecules are identical to the old molecule. One of the chains in each new molecule was present in the original molecule
This process = semi-conservative replication
Name two enzymes now known to be involved in DNA replication and explain their functions
DNA lipase; unwinds and unzips the DNA, breaking hydrogen bonds between the complementary base pairs and forming 2 separate strands
DNA polymerase; joins the new nucleotides to each other by phosphodiester bonds via condensation reactions forming the sugar-phophate backbone. Joins free nucelotides to the ends of the new DNA strands
DNA replication fork diagram
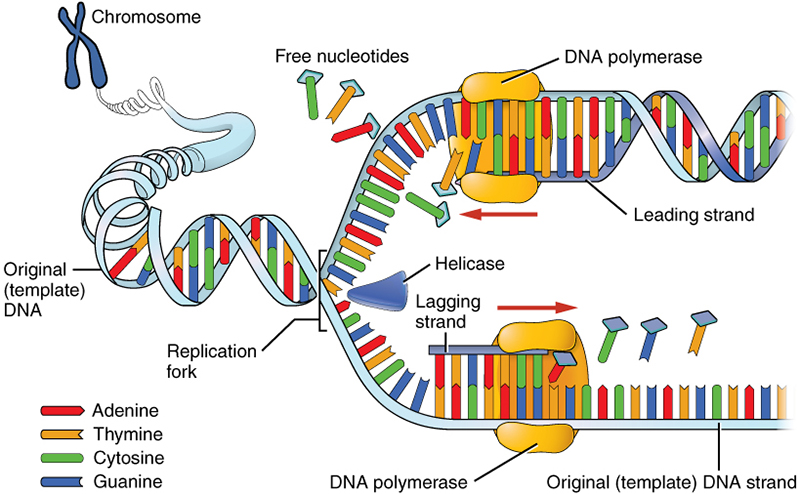
Direction of DNA replication
5’ to 3’
The genetic code
The genetic code is a linear, triplet, non-overlapping, degenerate, unambiguous, universal code for the production of polypeptides
The genetic code as linear
It is a linear code with no overlaps, so it can be ‘read’ without any ambiguity.
The genetic code as a triplet code
3 bases code for one amino acid
The genetic code as non-overlapping
Each base is part of only one triplet
The genetic code as degenerate
more than one triplet can specify an amino acid
The genetic code as unambiguous
each each triplet/codon specifies only one amino acid/stop codon
The genetic code as universal
is the same in all living things, so each specific codon codes for the same amino acid in all species
The triplet code for amino acids
3 bases = 1 amino acid
Amino acids are coded for by triplets of bases in the DNA.
The DNA is transcribed to produce codons in mRNA and then translated to produce a sequence of amino acids.
Exons
regions of the nucelotide sequence of DNA that contain the code for proteins
remains present in the final mature mRNA, after introns have been removed
Introns
Regions of non-coding DNA between exons
removed by post-transcriptional modification from pre-mRNA
Eukaryotic genes
usually discontinuous genes with coding exons and non-coding introns
Prokaryotic genes
usually continuous genes, lacking non-coding sequence.
Stages of protein synthesis
Transcription (DNA to mRNA)
Translation (mRNA to polypeptide)
Process of transcription
occurs in the nucleus
the molecule formed is called pre-mRNA as it contains both exons and introns
DNA helicase binds to DNA and unzips a section (gene) of the DNA by breaking the hydrogen bonds between complementary base pairs in the helix. This seperates the 2 polynucleotide strands by breaking the hydrogen bonds
The unpaired bases on the template strand are exposed
One chain acts as a tenplate for the formation of mRNA
RNA polymerase links to the template (coding) strand of DNA and attaches mRNA nucleotides one at a time to their complementary base pairs, e.g. Adenine in DNA now pairs with the mRNA base Uracil, Cytosine continues to pair with Guanine.
RNA polymerase joins the nucelotides via forming phosphodiester bonds between ribonucleoitdes, condensing the ribose-phosphate backbone
This copying stops at a stop sequence / codon. RNA polymerase detahces. mrRNA leaves the nucleus via the nucelar pores when a stop sequence is reached
The newly made premRNA then leaves the DNA.
Post-transcriptional modification of the pre-mRNA takes place to remove the non-coding introns, leaving only the coding sections (exons) in the mature mRNA. This leaves the nucleus to be translated into a protein in the cytoplasm.
Post-transcriptional modification
Post- transcriptional modification of pre-mRNA then occurs to remove the introns from the molecule to produce functional mRNA.
removal of introns (remain inside nucleus)
Exons are spliced together
Post-transcriptional modification in eukaryotes
in eukaryotes, mRNA contain coding (exons) and non-coding (introns) regions
Introns must be removed by splicing - a newly synthesised pre-mRNA is transformed into a mature mRNA - involves the removal of introns and joining of exons
Prokaryotic vs Eukaryotic Transcription
Prokaryoties; no post-transcription modification
Eukaryoties; the introns are removed by splicing
Process of Translation
mRNA is a linear chain of 3 base codons. There are complementary anticodons on tRNA molecules
The large subunits of a ribosome have 2 attachment sites for tRNA. The ribosome holds the mRNA and the tRNA (which have attached amino acids) in position for the amino acids to form peptide bonds and create a polypeptide chain. The codon on the mRNA (3 base code) therefore determines the tRNA as the tRNA which attaches must have a complementary 3 base code. E.g. mRNA CGA, tRNA GCU.
The tRNA that matches the codon on the mRNA has a specific amino acid attached to the 3’ end of the tRNA molecule.
Termination;
When the mRNA leaves the nucleus, it attaches to the small subunit binding sites of a ribosome.
Ribosome attaches to mRNA’s ribosome binding site and moves along to start codon
Two codons exposed on mRNA
First tRNA interacts with first codon and is attached to a specific amino acid via complemenentary base pairing
A second tRNA interacts with a second codon
The ribosome catalyses the formatio of s paptide bond between 2 amino acids, forming a dipeptide
The ribosome moves along the mRNA holding each tRNA in place until the amino acid attaches. In this way the mRNA (translated from a gene) carries the code for the formation of a polypeptide chain with amino acids set out in a particular order.
Elongation;
The first tRNA leaves the ribosome
Thr ribosome moves along the mRNA by 3 nucleotides, exposing another codon
The tRNA then leaves, the ribosome moves along and the next tRNA attaches to the next codon
Another tRNA interacts with the newly exposed codon
The ribosome catalyses the formation of a peptide bond, forming a polypeptide chain of 3 amino acids
Termination;
The sequence repeats until a ‘stop’ codon is reached
The mRNA seperates from the ribosome
The polypeptide sperates from the ribosome
In this way the mRNA (translated from a gene) carries the code for the formation of a polypeptide chain with amino acids set out in a particular order.
Prokaryotic vs Eukaryotic Translation
Prokaryotic; less post-translational modification, Chemical modification
Eukaryotes; the start of the protein may be removed and carbohydrates added to form glyco-proteins
Stop codon
A sequence of 3 nucelotides in DNA or mRNA that signals a halt to protein synthesis in the cells
Gene
A section or sequence of DNA that codes for a polypeptide
Explain the steps in the process that produces viralm proteins from viral mRNA, using host cell machinery. [5]
The viral mRNA is translated to produce a viral protein
Ribosomes attaches, exposing the codons on the mRNA
tRNA with complementary anticodons interact with exposed codons
the tRNA are each attached to a single specific amino acid
The ribosome forms a peptide bond betweeen the 2 amino acids
The ribosome moves along 3 nucleotides and the process repeats
When the ribosome reaches a stop codon, it detaches
The ‘one gene - one polypeptide’ hypothesis
each gene is transcribed and then translated into a single polypeptide
The further modification and combination of some polypeptides
May be structurally or chemically modified = post-translational modification
Polypeptides can be chemically modified in the Golgi Body by combination with non-proteins such as ;
+ carbohydrates —> glycoproteins
+ lipids —> lipoproteins
+ phosphate —> phospho-proteins
Structural modification; in the ER, many polypeptides are folded into secondary, tertiary or quaternary structures. occasionally the primary structure of a polypeptide is functional but it is usually folded/strtucturally modified
Polypeptides can be combined, as exmeplifies by haemoglobin; highly-modified molecule. each polypeptide has an alpha-helix region (secondary structure), and is folded (tertiary structure). 4 polypeptides are combined (quaternary structure). the protein is modified by combination with 4 non-protein haem groups to make the functional molecule
Draw a simple labelled diagram to show the structure of a nucelotide
What are the components of ATP?
3 phosphate groups with high energy bonds between them
ribose (pentose sugar)
adenine (nitrogeous base)
What is the difference between an endergonic and exergonic reaction?
Endergonic reaction =a reaction that requires an input of energy. e.g. ATP synthesis via condensation
Exergonic reaction = a reaction that releases energy. e.g. ATP hydrolysis. The energy required to combine ADP and inorganic phosphate (Pi) to form ATP (and water) comes from exergonic reactions e.g. cell respiration.
How much energy is released from one mole of ATP when it is hydrolysed to ADP and Pi?
30.6kJmol^-1
What does universal energy currency mean?
common energy source used in all living organisms
List 3 advantages of using ATP to provide energy in cells
ATP releases energy in one hydrolysis reaction controlled by one enzyme.
ATP releases energy in small, usable amounts. ATP travels easily to where it may be used for secretion, muscle contraction, nerve transmission or active transport.
What is the sugar in DNA?
deoxyribose
Which bases are in the nucleotides of DNA?
Cytosine and Guanine, Adenine and Thymine
What is the sugar in RNA?
Ribose
What are the bases in RNA?
Cytosine and Guanine, Adenine and Uracil
How does the structure of ATP differ from that of DNA?
ATP;
3 phosphate groups bonded together by high energy bonds
ribose (pentose sugar)
adenine (nitrogenous base)
DNA;
1 phosphate group
deoxyribose
Adenine, Thymine, Cytosine or Guanine
How does the structure of ATP differ from that of RNA?
ATP;
3 phosphate groups bonded together by high energy bonds
Ribose
Adenine
RNA:
1 phosphate groups
Ribose
Adenine, Uracil, Cytosine or Gunaine
What does DNA stand for?
Deoxyribonucleic acid
What does RNA stand for?
Ribonucleic acid
What are the 2 functions of DNA?
replication and protein synthesis
What are the 3 types of RNA?
mRNA (Messenger RNA)
rRNA (Ribosomal RNA)
tRNA (Transfer RNA)
Which is longer; DNA or RNA?
DNA as it contains all of the complete genetic information of an organism
tRNA and rRNA are short; mRNA varies but shorter than DNA
Which bases are pyrimidines and which are purines?
Pyrimidines; Thymine, Cytosine and Uracil
Purines; Adenine and Guanine
Complementary bases are linked by bonds, what type of bonds are these?
Hydrogen bonds
2 between C and G
2 between A and T/U
What does antiparallel mean?
Running parallel but facing in opposite direction
What is the shape of DNA?
Double Helix
Why does DNA replication need to occur?
When cells divide to form new daughter cells, they must receive an exact copy of the DNA. Therefore, chromosomes must be able to make exact copies of themselves.
What is the accpeted model of DNA replication?
Semi-conservative replication
What does semi-conservative replication mean?
Each of the strands of DNA acts as a template to make a new strand. New DNA molcules are made of an original strand linked to a new strand
Meselson and Stahl carried out an experiment which gave evidence to the theory of semi-conservative replication of DNA.
What are the main steps in DNA replication?
Replication starts at a specific sequence on the DNA molecule; the replication origin
DNA helicase unwinds and unzips DNA, breaking the hydrogen bonds that join the complementary base pairs, forming 2 separate strands
Bases are now exposed and unpaired
Free nucleotides in the nucleoplasm are bound to their complementary bases on the unzipped strand.
DNA polymerase joins the nucleotides together by condensation reactions between sugar and phosphate groups of adjacent nucleotides, forming phosphodiester bonds in the new strands of DNA being formed
A winding enzyme winds the new strands up to form double helices
2 new DNA molecules are formed from 1 old molecule and 1 new molecule. The 2 new molecules are identical to the old molecule. One of the chains in each new molecule was present in the original molecule
This process = semi-conservative replication
What is the enzyme that catalyses DNA replication?
DNA helicase, then DNA polymerase
Which 2 scientists undertook the experiment that showed how DNA replication happens?
Meselson and Stahl
Which part of the DNA molecule was labelled in the Meselson and Stahl experiment? What were they labelled with?
Nitrogenous bases with isotopes of nitrogen
Draw the results for generation 0, 1 and 2 of the Meselson and Stahl experiment, labelling the lines on the tubes
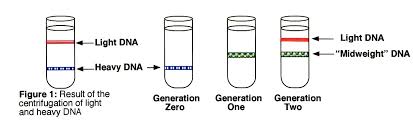
How do the results of the Meselson and Stahl experiment show that conservative and dispersive replication do not occur?
Meselsohn and Stahl’s experiment disproves alternative theories.
Conservative replication – direct copying of the nucleotide sequence onto a new double stranded molecule which would give one light and one heavy molecule in generation 1.
Dispersive replication – where half the nucleotides are placed randomly in the DNA being replicated to make new molecules which would give successively lighter molecules and therefore a band between hybrid and light in generation 2
What does DNA polymerase do?
joins the new nucleotides to each other by phosphodiester bonds via condensation reactions forming the sugar-phophate backbone. Joins free nucelotides to the ends of the new DNA strands
What does the base sequence in DNA code for?
The order of amino acids in a protein
How many bases code for each amino acid?
3
What is the process by which the code is copied from DNA to mRNA?
Transcription in the nucleus
What is the relationship between DNA and mRNA?
mRNA is transcribed from DNA
Which enzyme controls transcription?
DNA helicase, then DNA polymerase
What is an exon and what is an intron?
Exon; regions of the nucelotide sequence of DNA that contain the code for proteins. remains present in the final mature mRNA, after introns have been removed
Intron; Regions of non-coding DNA between exons. removed by post-transcriptional modification from pre-mRNA
What is meant by post-transcriptional modification?
Post- transcriptional modification of pre-mRNA then occurs to remove the introns from the molecule to produce functional mRNA.
removal of introns (remain inside nucleus)
Exons are spliced together
Where are proteins synthesised?
Ribosomes in cytoplasm and RER
How does mRNA leave the nucleus?
Via the nuclear pore
State 2 structural differences between DNA and RNA
DNA has a deoxyribose sugar. RNA has a ribose sugar
DNA is a double-stranded double helix. RNA is single stranded