OCR A Level Biology Module 6
1/231
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
232 Terms
regulatory gene
a gene that codes for a transcription factor, so controls the expression of structural genes
structural gene
gene that codes for polypeptide
mutation
a random change in the sequence of bases in an individual gene, or in the structure of the chromosome (which changes the sequence of genes on the chromosome)
examples of mutagenic agents
x-rays, UV light (physical), nitrous acid (chemical) & some viruses (biological agents)
what can chromosomal mutations also do?
alter the chromosome no of the individual
what can cause chromosomal mutations?
- deletion
- inversion
- translocation
- duplication
- non-disjunctionexa
what are the 3 types of substitution mutations?
- silent - there's a change in the base sequence, but it still codes for the same aa
- missense - a change to the base sequence that leads to a change in the aa sequence in a protein
- nonsense - a point mutation may alter a base triplet, so it becomes a stop codon. This stops transcription and results in a truncated protein
how can a mutation in a gene lead to a truncated protein?
1. A mutation in the DNA can lead to a stop codon
2. translation is terminated by the stop codon
3. no more aas are added after the stop codon
how can a mutation in a gene lead to a non-functioning enzyme?
1. there is a change in the base sequence of the gene, leading to a change in the aa sequence of an enzyme
2. this results in a change in the hydrogen, ionic, or disulphide bonds
3. leading to a change in the tertiary structure, and the shape of the active site in the enzyme
4. so the substrate is no longer complementary
5. and can't bind to the active site so no E-S complexes form
examples of gene mutations
sickle cell anaemia, cystic fibrosis, albinoism, Down's Syndrome
characteristics of Down's Syndrome
- an individual receives 3 copies of chromosome 21 instead of 2 due to translocation of a chromosome
- people with Down's Syndrome have learning difficulties & thick set bodies
transcription factors
proteins, or short non-coding RNA, that can combine with a specific site on a length of DNA and inhibit or activate transcription of the gene
what does E. coli usually use as a respiratory substrate?
glucose
what does E. coli use when this is absent?
lactose, because it induces the production of the enzymes lactose permease, which allows lactose to enter the bacterial cell, and B-galactosidase, which hydrolyses lactose to glucose & galactose
what happens in the lac operon in the presence of lactose?
1. mRNA is transcribed by the regulatory gene
2. the repressor protein is translated from the mRNA strand
3. lactose binds to the repressor protein & changes its shape
4. this stops it binding to the operator region
5. so RNA polymerase binds to the promoter region
6. the structural genes Z & Y are transcribed into mRNA
7. the mRNA is translated into lactose permease & B-galactosidase
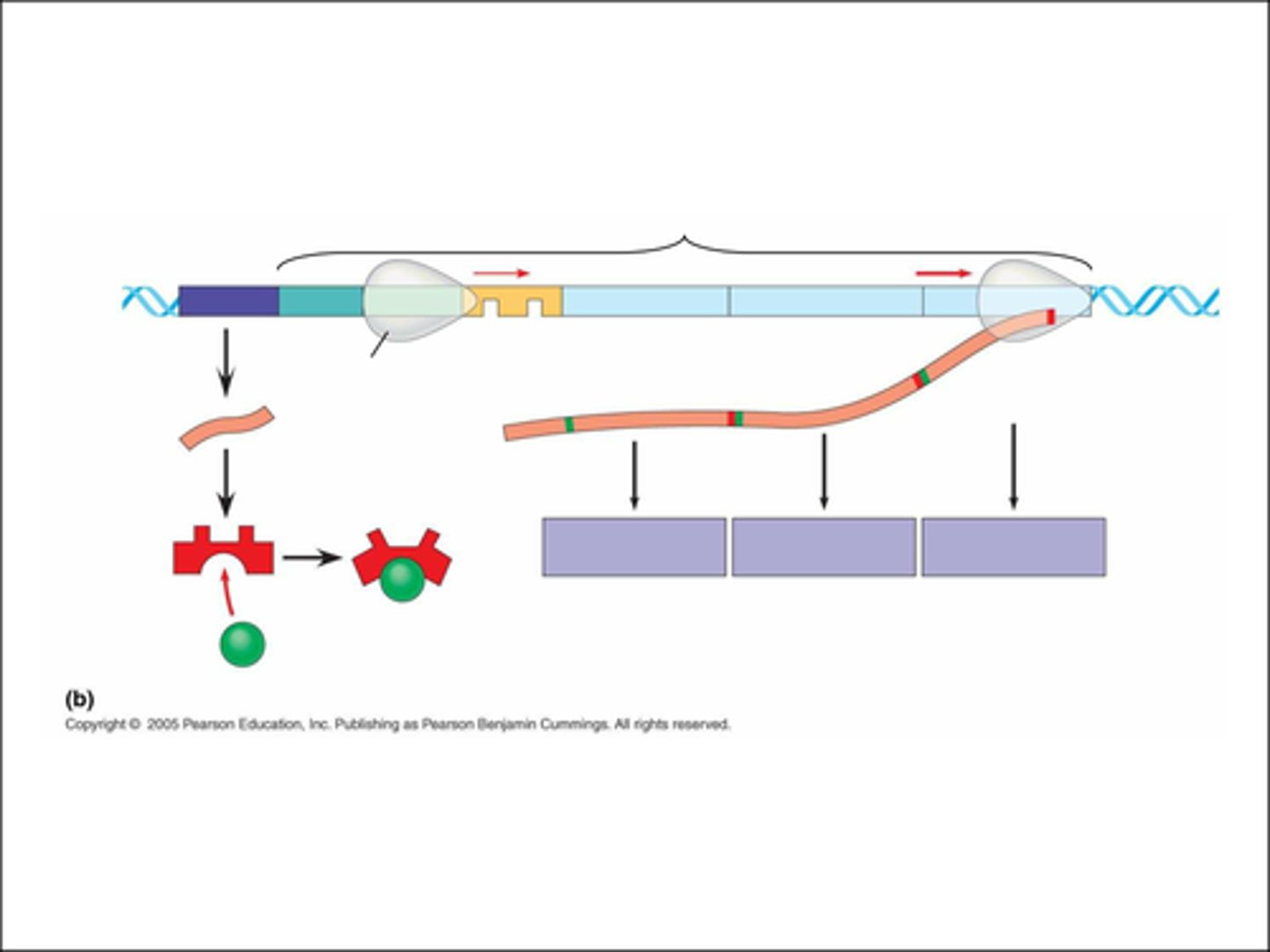
at what level does the lac operon control gene expression?
transcriptional level
regulatory mechanisms that control gene expression at post-transcriptional level
- primary mRNA is modified
- introns are removed to produce mature mRNA
- alternative splicing can produce different versions of mRNA
regulatory mechanisms that control gene expression at post-translational level
- proteins must be activated by cAMP by phosphorylation
- the binding of cAMP alters the shape of the protein
-it alters the 3D structure of the protein by phosphorylation, in which a Pi group is attached to the protein
homeobox gene
a homeotic gene that contains a homeobox sequence consisting of 180 base pairs that codes for a homeodomain sequence
what does the homeodomain sequence do?
binds to DNA & acts as a transcription factor. It initiates transcription and is involved in the control of development & the body plan
Why are homeobox genes similar in plants, animals and fungi?
- there has been very little change by mutation in homeobox genes because these genes are very important & mutation would alter the body plan
- many other genes would be affected so the mutation would be likely to be lethal
hox genes
a subset of homeobox genes, found only in animals; involved in formation of anatomical features in the correct locations of the body plan.
apoptosis
programmed cell death
the process of apoptosis
1. enzymes break down the cell cytoskeleton
2. the cytoplasm becomes dense with tightly packed organelles
3. blebs form from the plasma membrane
4. chromatin condenses, the nuclear envelope breaks & DNA breaks into fragments
5. the cell breaks into vesicles that are removed by phagocytosis
6. so the cell debris isn't released into surrounding tissue
at what level do hox genes & apoptosis control gene expression?
development
why can failure of the control mechanisms during development lead to deformities?
- it can stop apoptosis occurring
- hox gene doesn't produce the transcription factor
- molecules signalling apoptosis aren't produced
how can meiosis lead to genetic variation?
- independent assortment of homologous chromosomes in metaphase i
- independent assortment of chromatids in metaphase i
- so homologous chromosomes come from different parents. This produces a large number of allele combinations
- crossing over in prophase i, so chromatids will have a new combination of alleles
- random mutations change the base sequence
- non-disjunction, where chromosomes do not separate in metaphase i
- random fertilisation of gametes
- gametes are not genetically identical
example of the environment interacting with genes
chlorosis in plants, which occurs when leaves don't develop enough chlorophyll if the soil plants are grown in doesn't contain enough magnesium
homologous chromosomes
a pair of 1 maternal & 1 paternal chromosome, that are the same length and have genes at the same loci
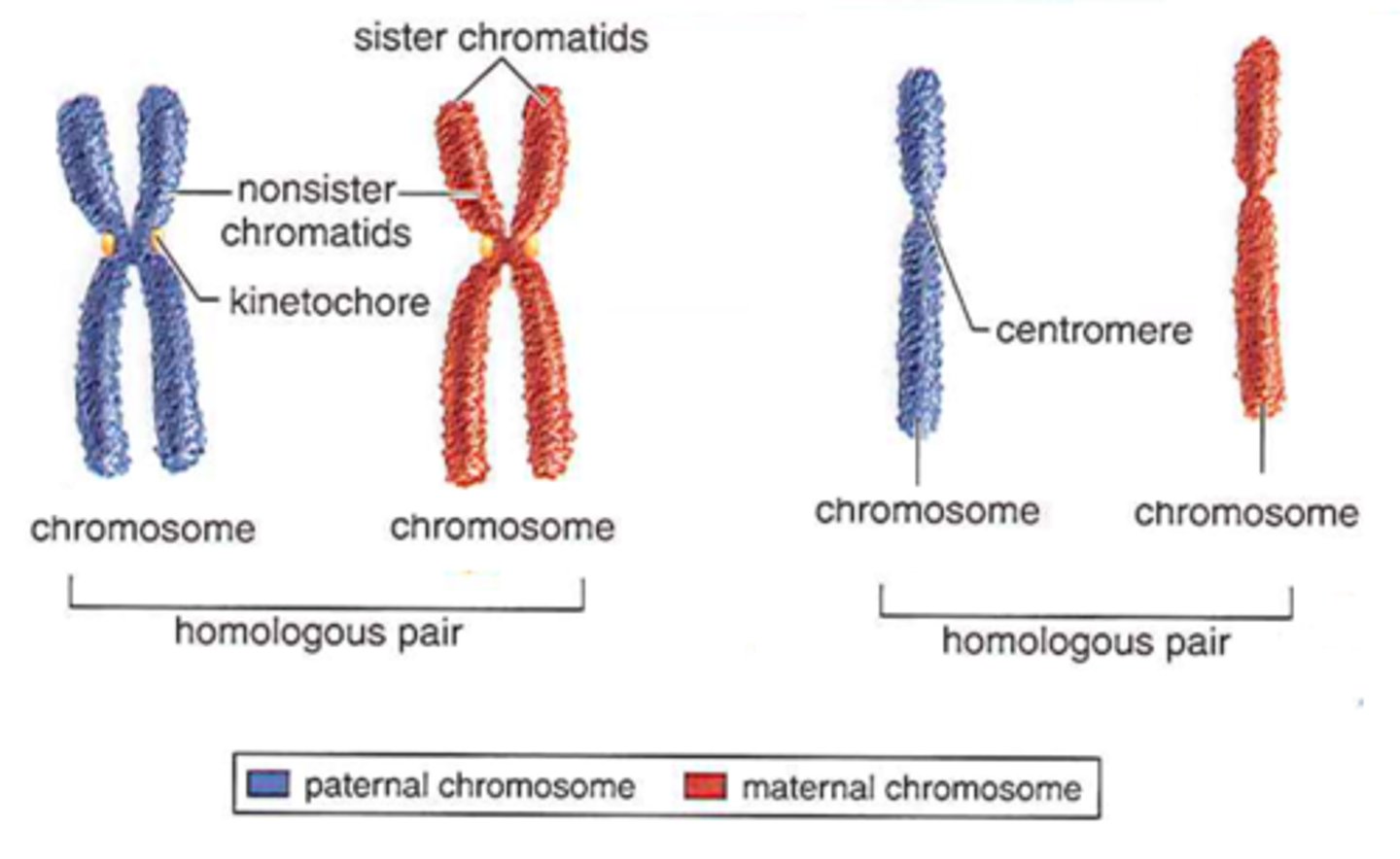
sister chromatids
the 2 identical chromatids produced when a chromosome has replicated
gene
a sequence of bases that codes for a polypeptide
locus
position of a gene on a chromosome
allele
a version of a gene.
phenotype
observable characteristics of an organism
genotype
alleles present within the cells of an individual for a particular characteristic
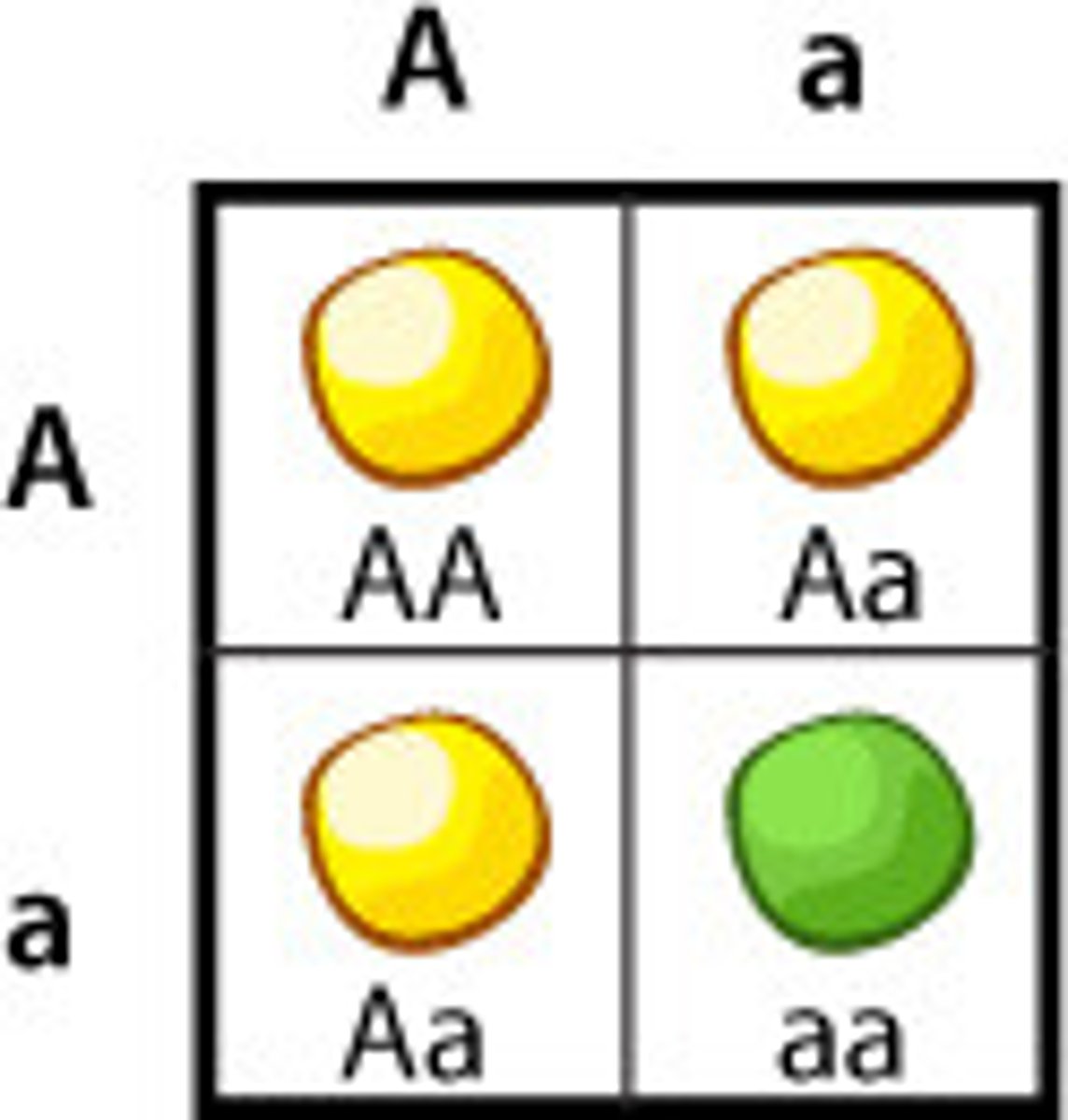
dominant
a characteristic in which the allele responsible is expressed in the phenotype, even in those with heterozygous genotypes
codominant
a characteristic where both alleles present in the genotype contribute to the phenotype.
recessive
a characteristic in which the allele responsible is only expressed in the phenotype if there is no dominant allele present
linkage
when genes for different characteristics that are present at different loci on the same chromosome are linked
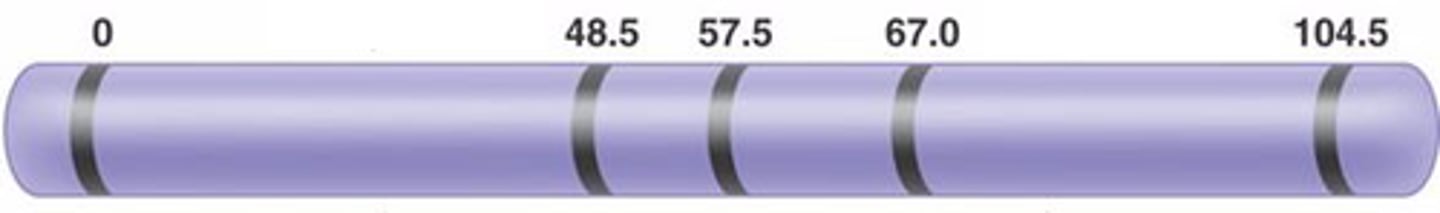
sex linkage
the presence of a gene on 1 of the sex chromosomes
autosomal linkage
gene loci present on the same autosome that are often inherited together
autosome
any chromosome that is not a sex chromosome
how to ensure that the offspring produced when 1 pure-bred plant is crossed with another are only from the desired cross
1. transfer the pollen to the stigma by hand using a paintbrush
2. isolate the plants to prevent pollination by foreign plants
3. bag the plants before & after pollination
4. remove the anthers before maturity using scissors, to prevent self-pollination
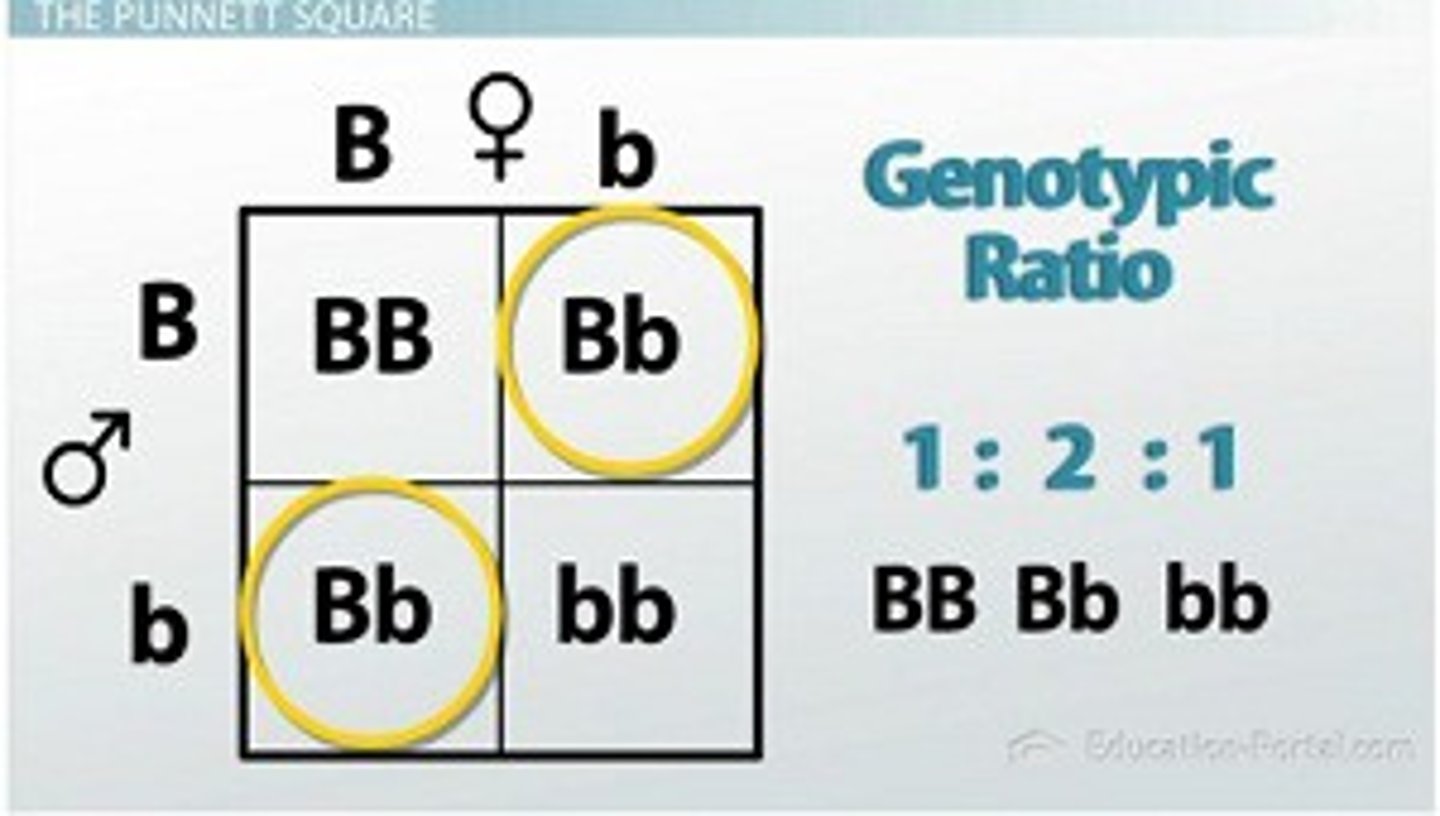
monogenic inheritance
when a phenotype is controlled by a single gene
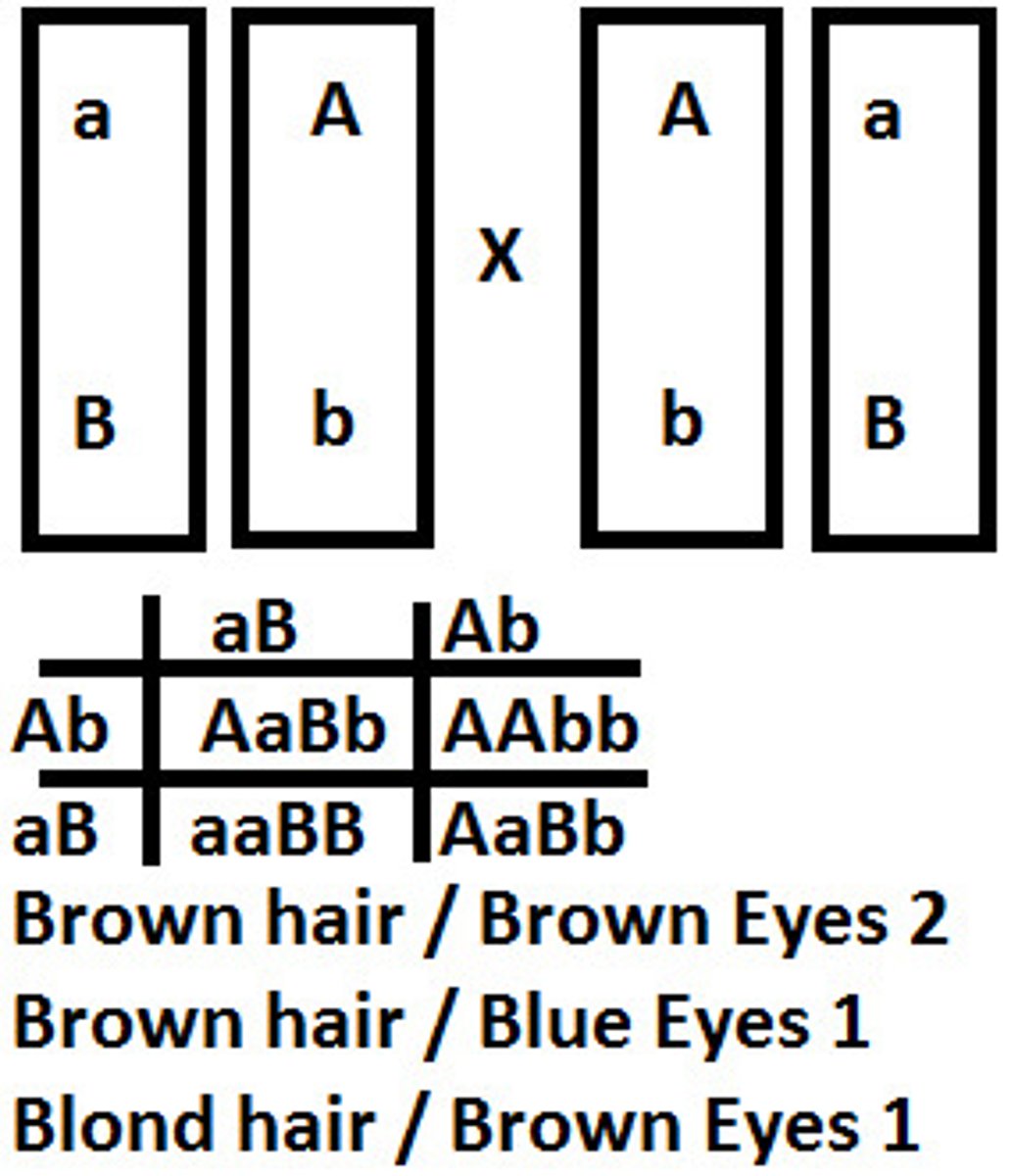
dihybrid cross
the inheritance of 2 genes / characteristics
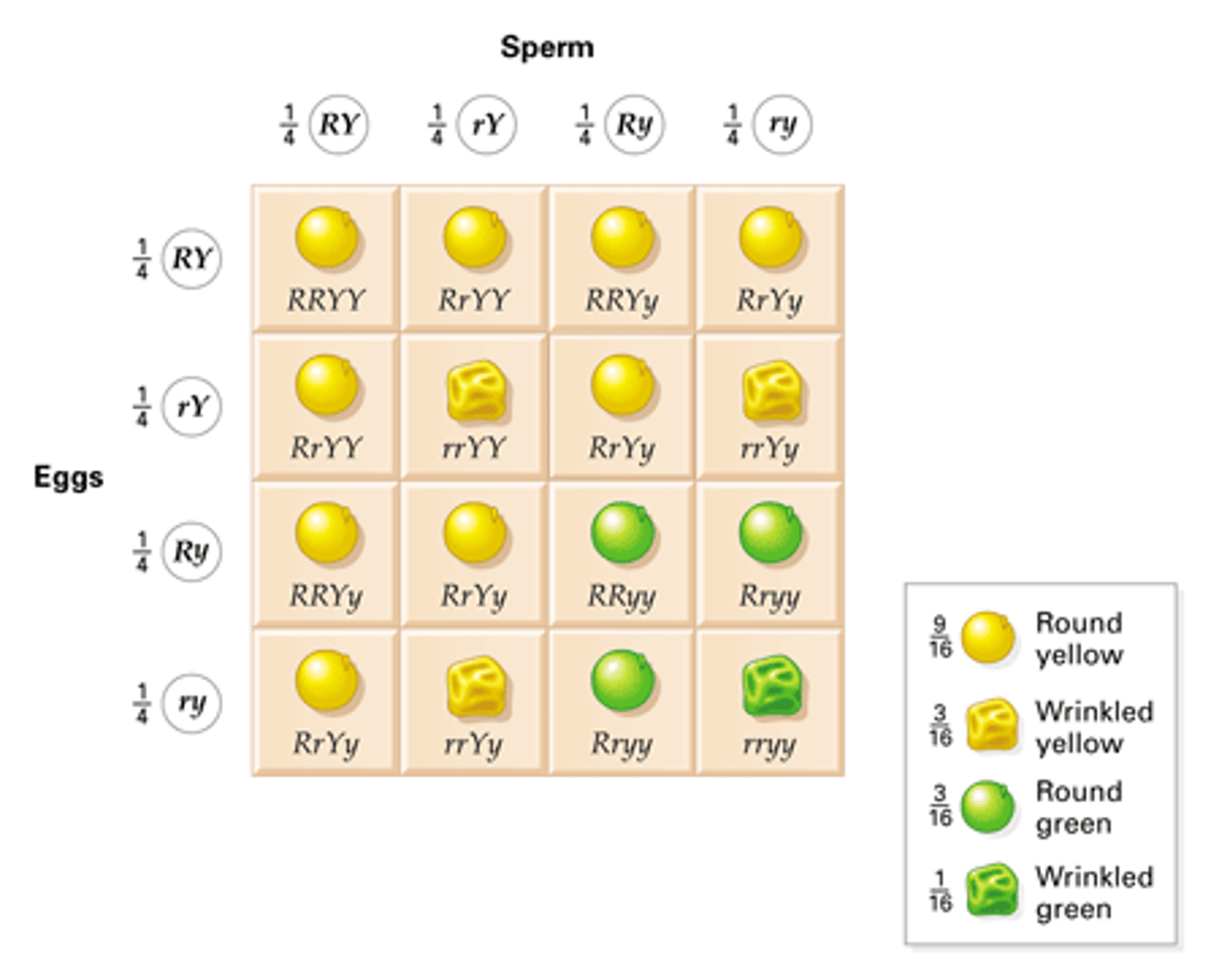
epistasis
the interaction of different gene loci, so that one gene masks / suppresses expression of another gene locus
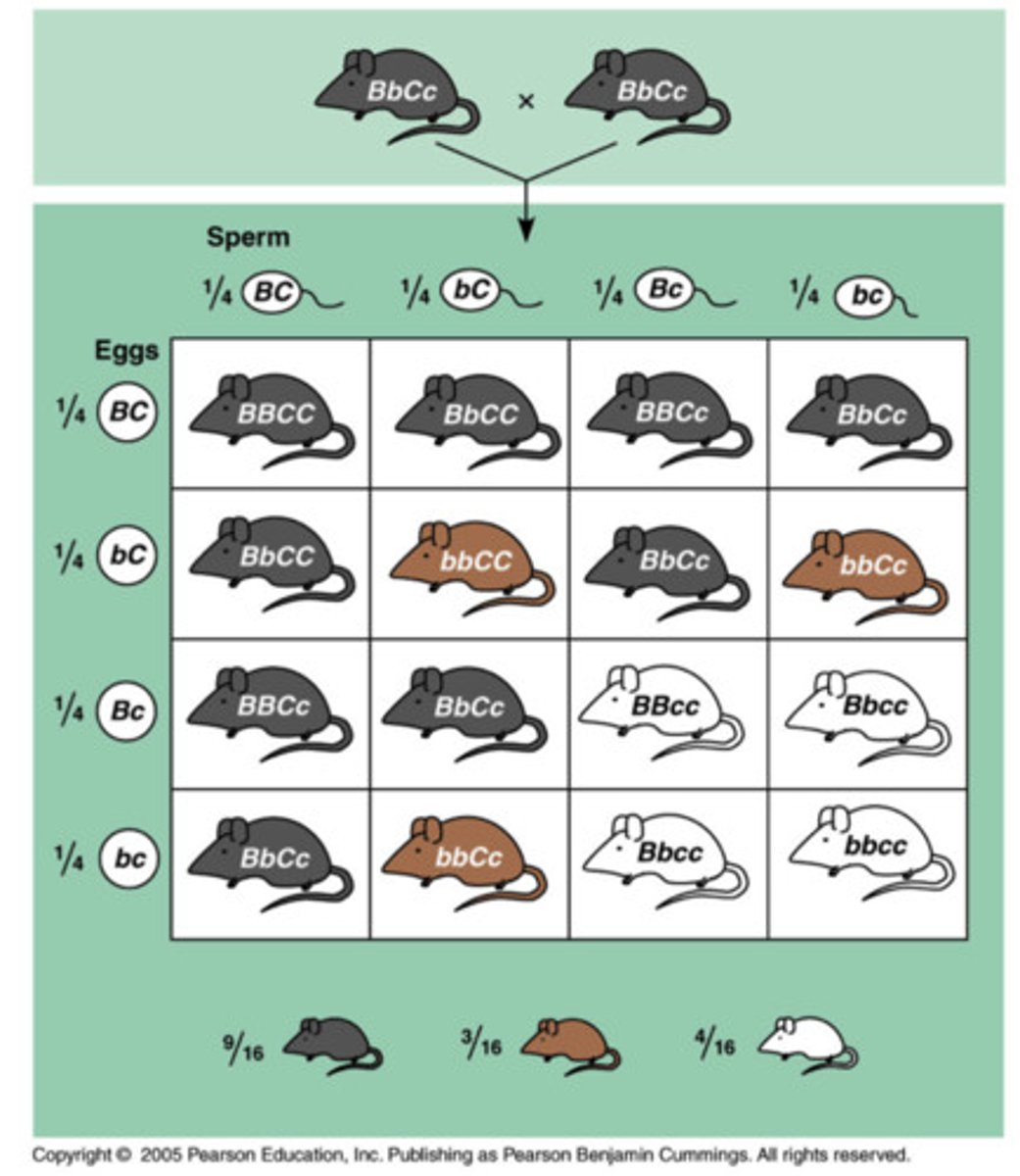
recessive epistasis
when the homozygous recessive allele of one gene is dominant to & masks the expression of either allele of the second gene at the second linked locus
recessive epistasis inheritance ratio
9:3:4
dominant epistasis
when the dominant allele of one gene masks the expression of all alleles of the second gene at the second linked locus
dominant epistasis inheritance ratio
12:3:1 or 13:3
complementary epistasis
when the homozygous recessive allele of one gene is dominant to & masks the expression of the dominant allele of the second gene at the second linked locus
complementary epistasis inheritance ratio
9:7
what does complementary epistasis involve?
one gene forms an intermediate compound
the other gene codes for an enzyme that converts the intermediate compound to the final pigment
how might 1 allele inhibit the expression of another?
1. allele 1 could code for a repressor protein or transcription factor
2. protein (product of allele 1) binds to promoter region of allele 2
3. & stops the transcription of mRNA so stops the translation of allele 2
chi-squared test
statistical test to find out if the difference between observed & expected data is significant or due to chance
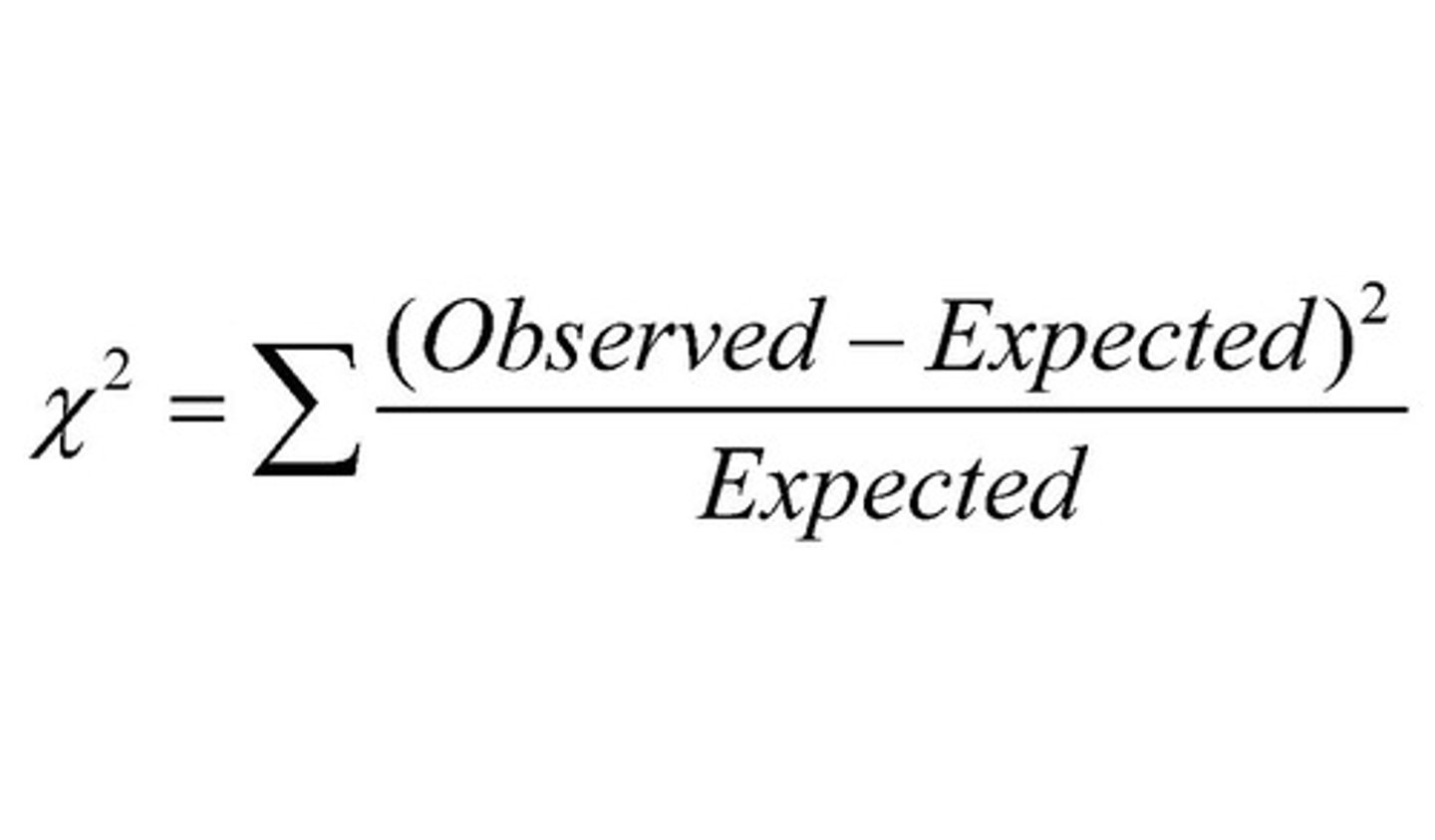
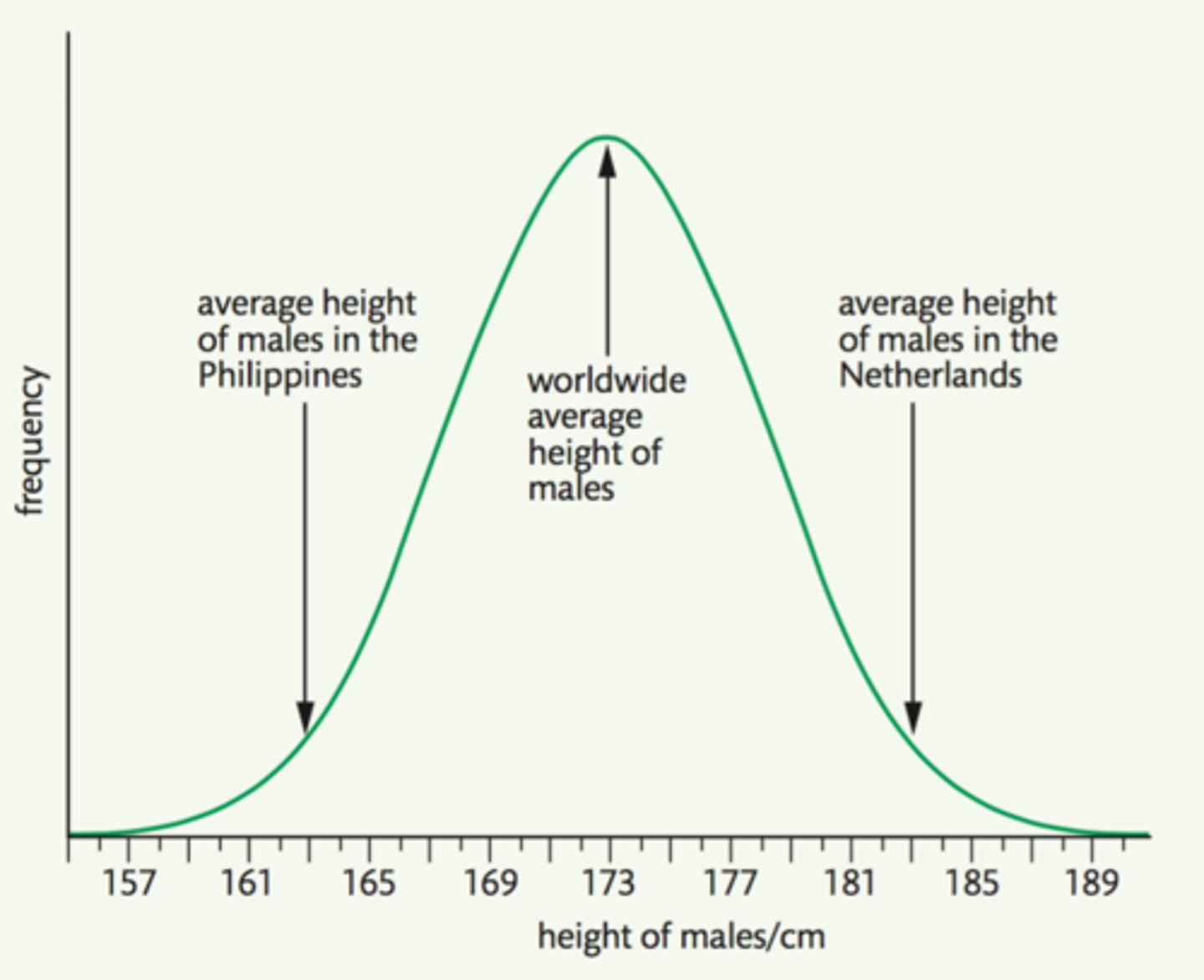
continuous variation
variation that produces phenotypic variation where quantitative traits vary by very small amounts between one group & the next
discontinuous variation
genetic variation producing discrete phenotypes - 2 or more non-overlapping categories
how many genes are involved in characteristics that exhibit discontinuous variation?
characteristics that exhibit discontinuous variation are usually determined by the alleles of a single gene locus, so they're monogenic
how many genes are involved in characteristics that exhibit continuous variation?
characteristics that exhibit continuous variation are usually determined by many genes, so they're polygenic
types of natural selection
directional, stabilising, disruptive / dispersive
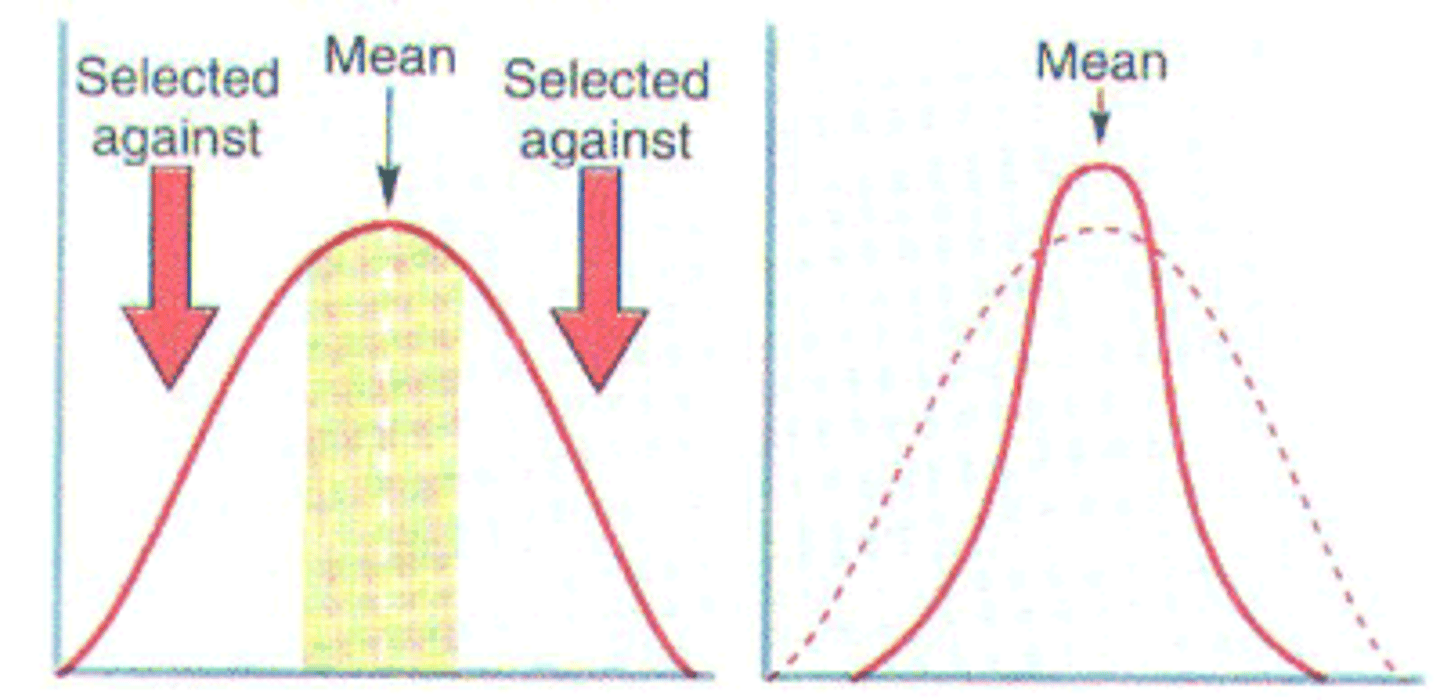
stabilising selection
natural selection that favours intermediate phenotypes, and extreme phenotypes are selected against. Stabilising selection decreases genetic variation within a population
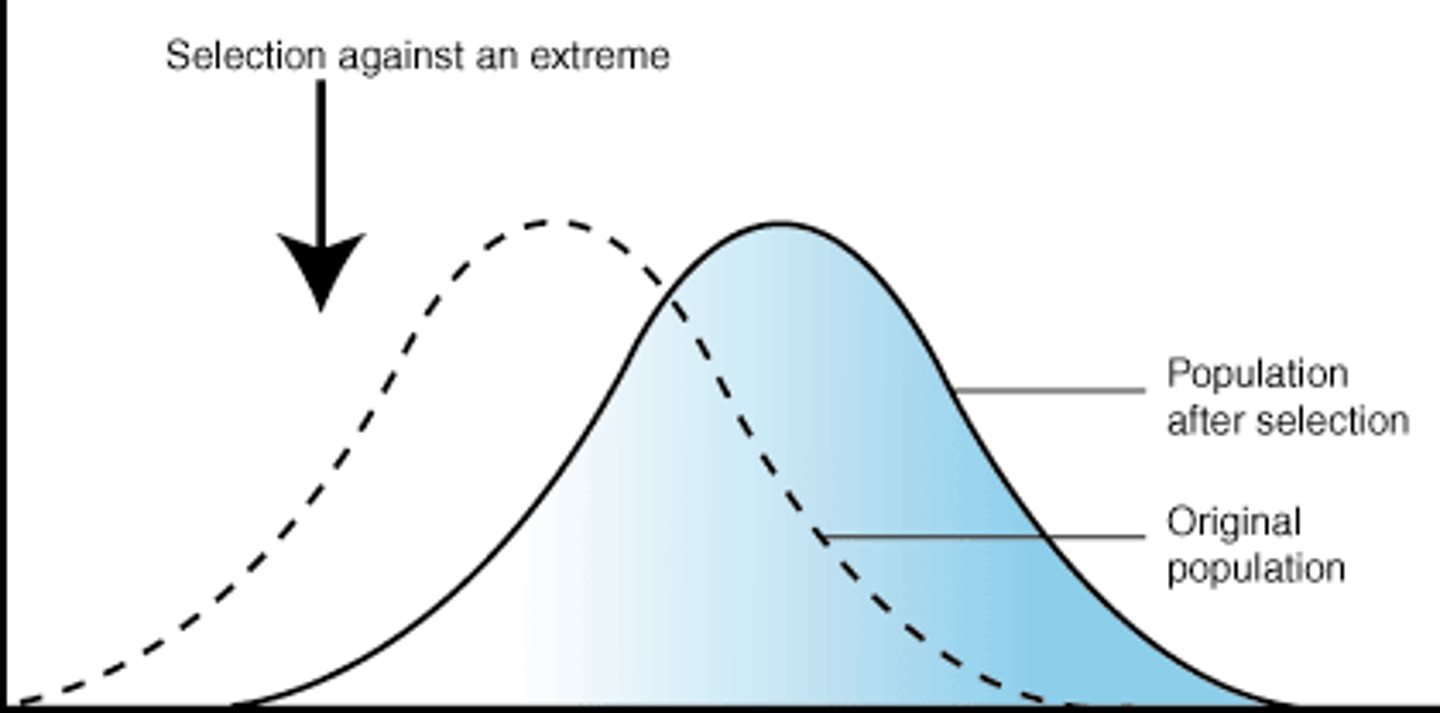
directional selection
natural selection that favours less common, more extreme phenotypes. This is because an environmental change favours a new phenotype & results in a change in the population mean
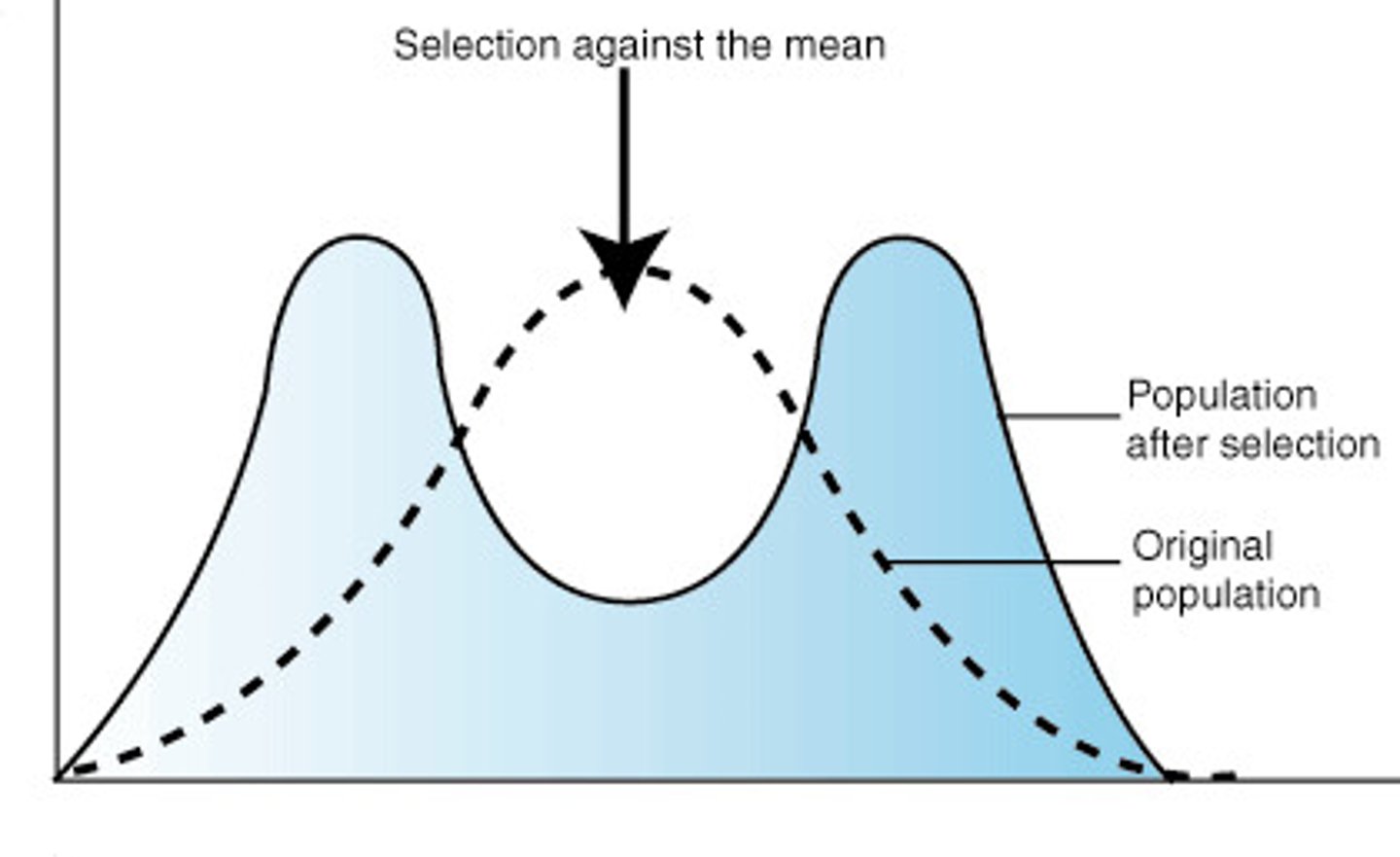
disruptive selection
natural selection that favours individuals at both extremes of the phenotypic range. It removes individuals from the centre of the phenotype
genetic drift
A change in the allele frequency of a population, which causes some phenotypic traits to become more or less common. It is a result of chance events rather than natural selection.
causes of genetic drift
genetic bottleneck effect and founder effect
genetic bottleneck
there's a sharp decrease in the size of a population due to an environmental catastrophe (e.g. an earthquake) which decreases genetic diversity. As the population increases again it is less genetically diverse than before
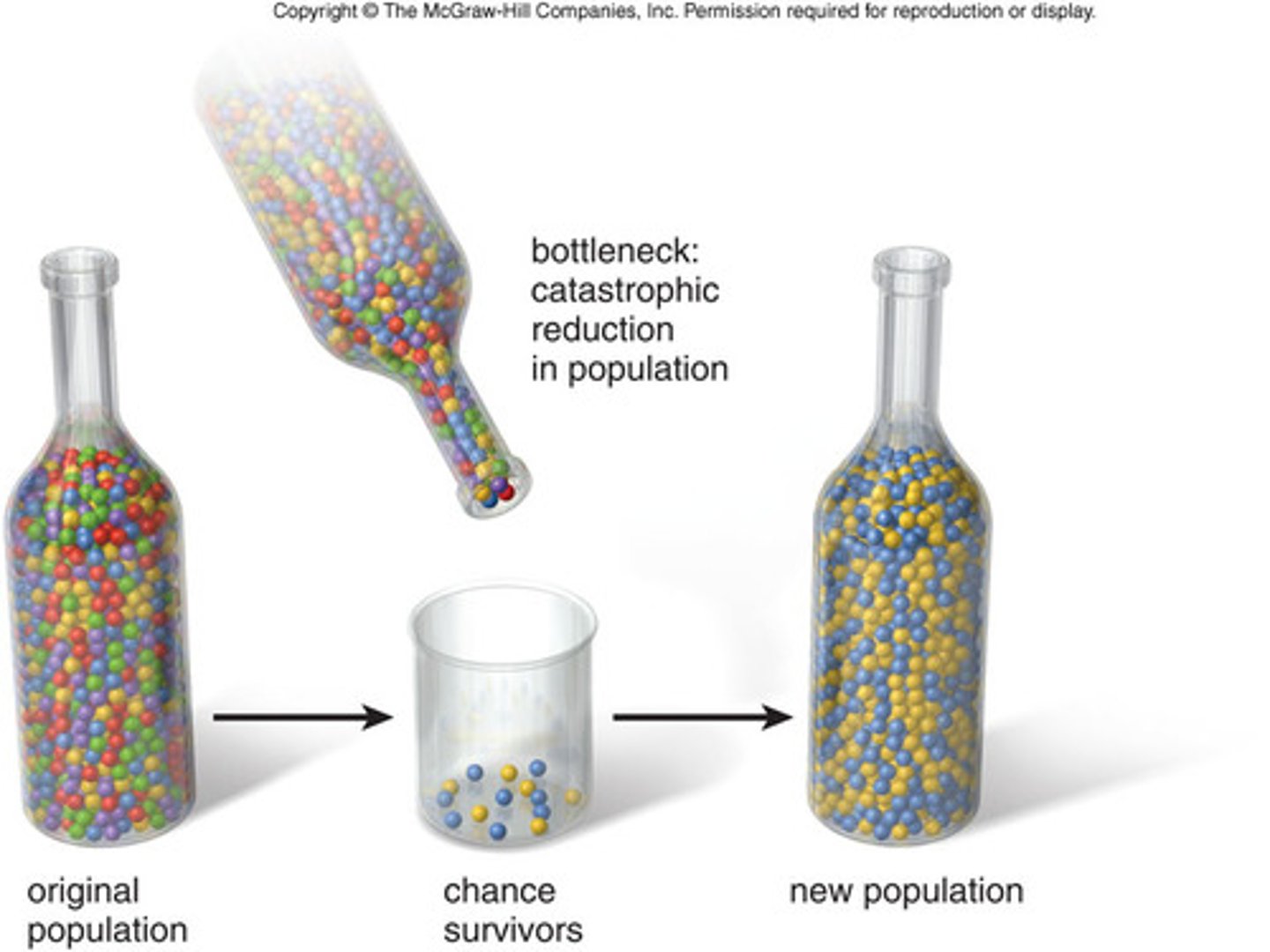
the founder effect
When a small sample of an original population establishes in a new area, its gene pool isn't as diverse as that of the parent population
the equations involved in the Hardy-Weinberg Principle
p = frequency of dominant allele in a population (B)
q= frequency of recessive allele in a population (b)
p2 = frequency of dominant genotype (BB)
q2 = frequency of recessive phenotype (bb)
2pq = the frequency of the heterozygous genotype (Bb)
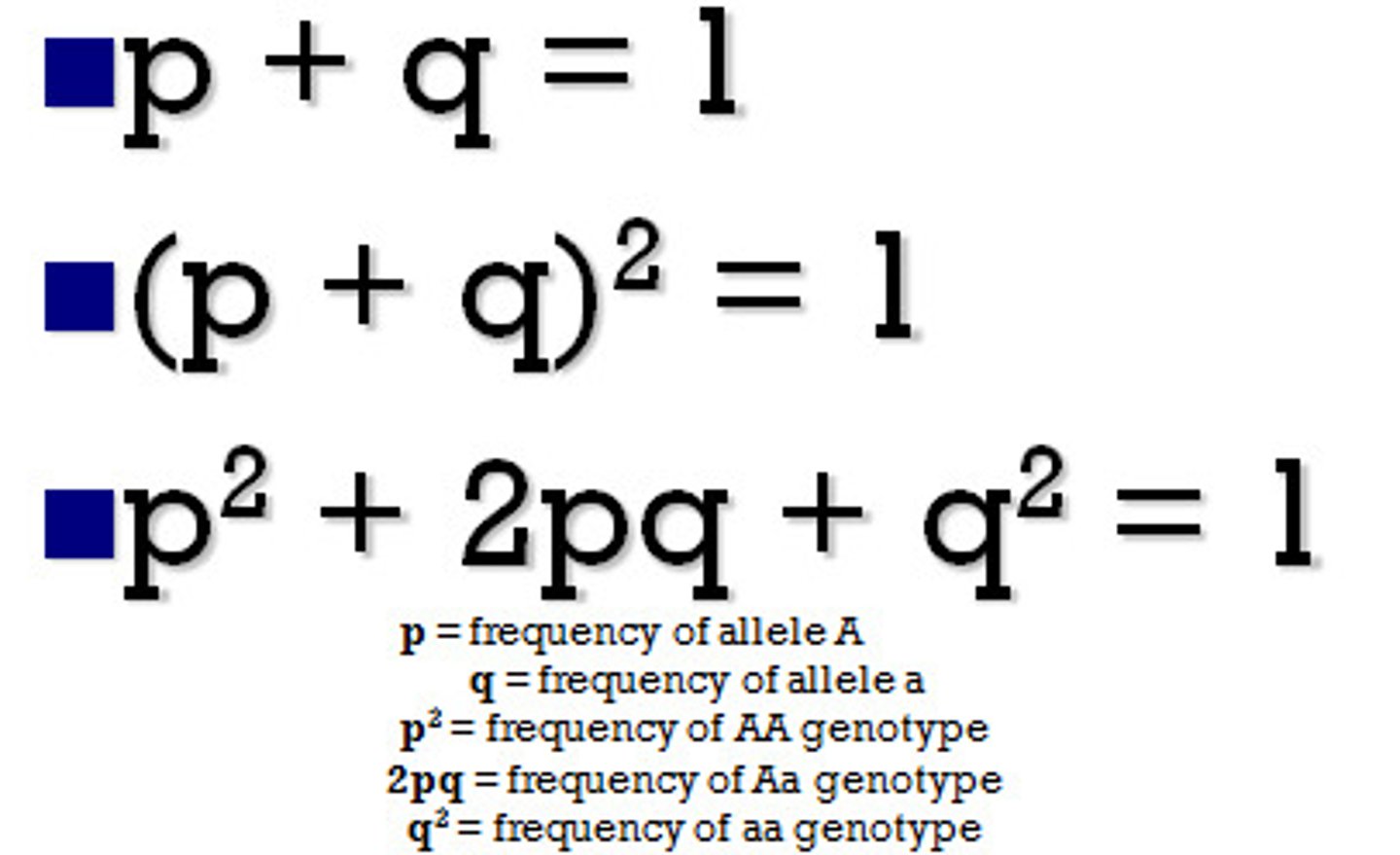
speciation
the splitting of a genetically similar population into 2 or more populations that undergo genetic differentiation & eventually reproductive isolation, leading to the evolution of 2 or more new species
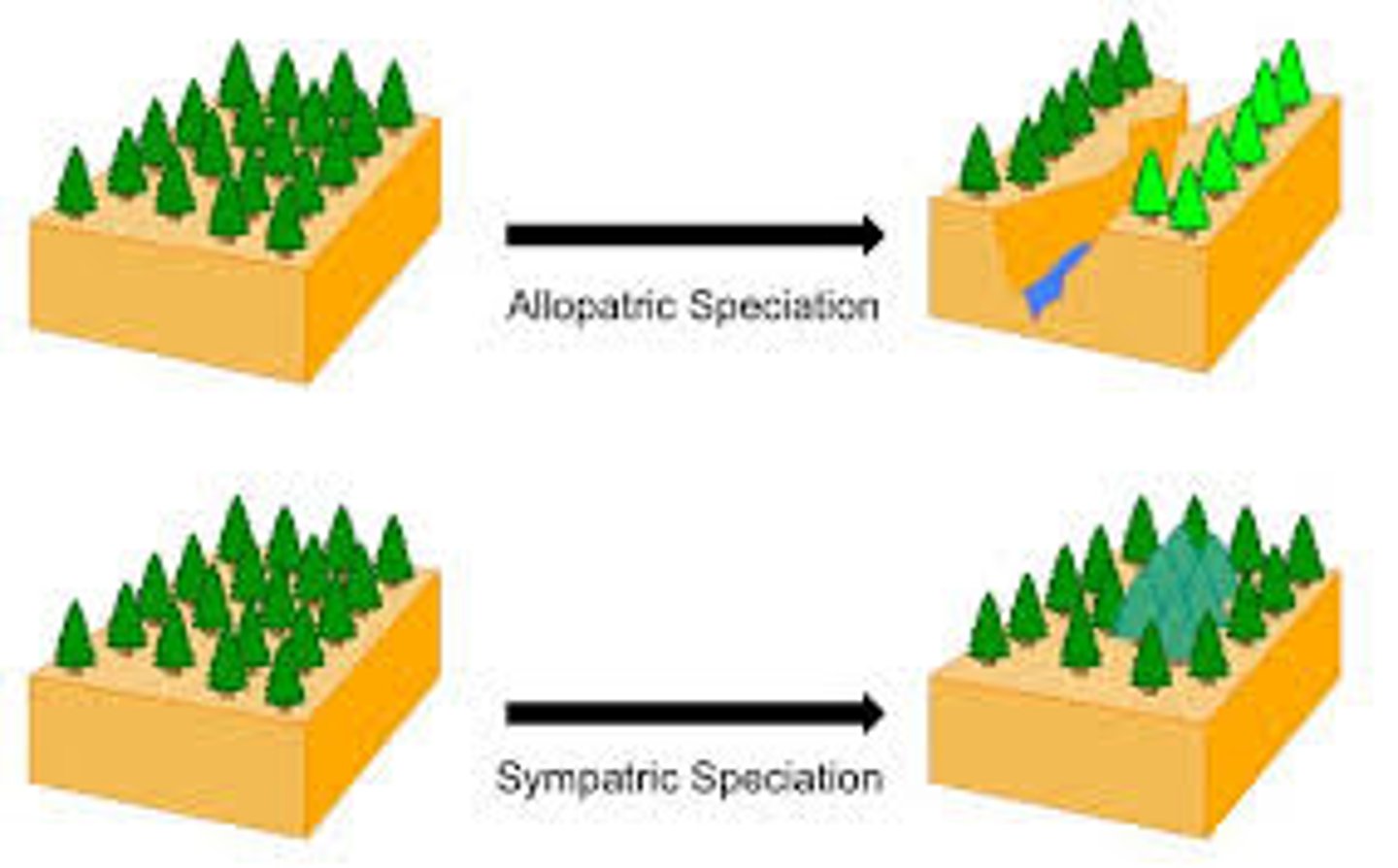
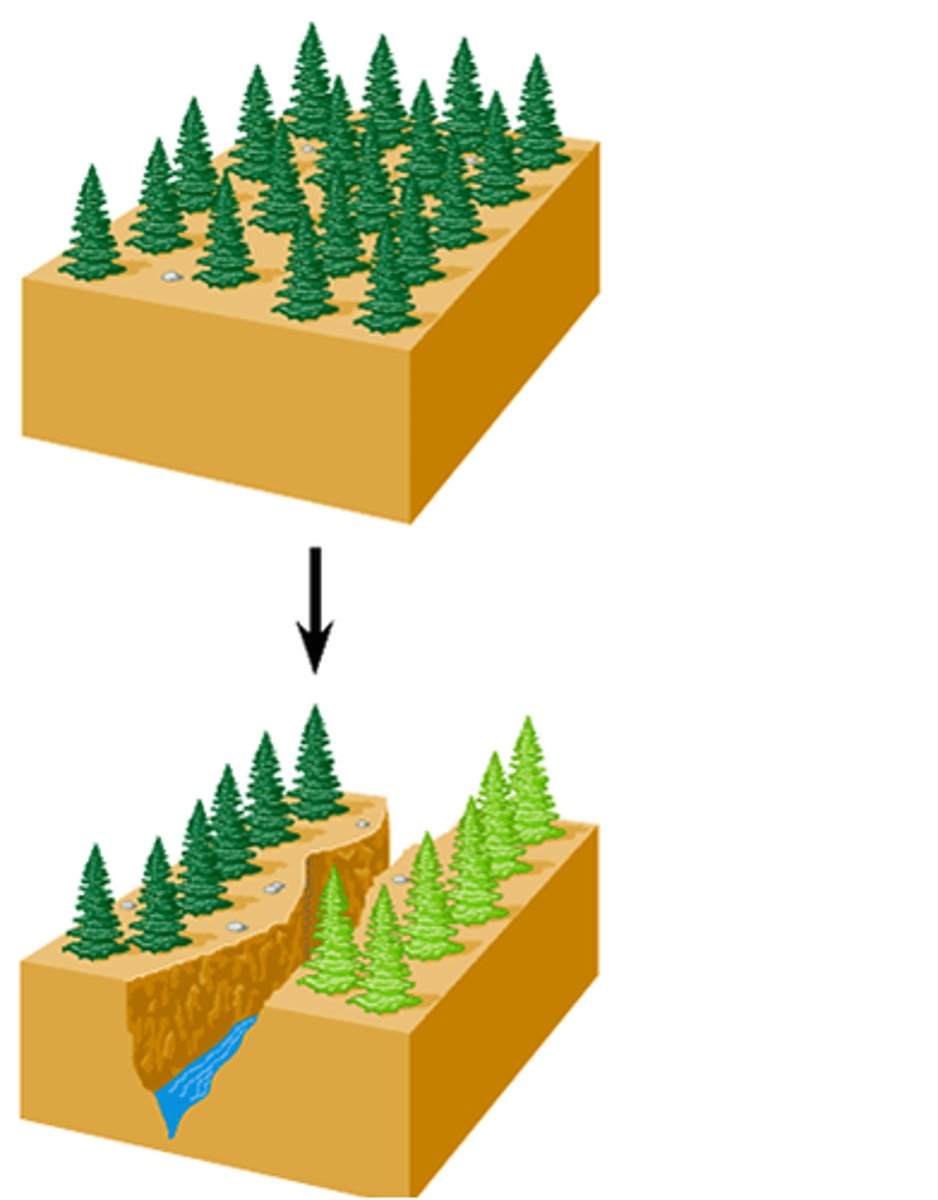
allopatric speciation
formation of 2 different species from 1 original species due to geographical isolation
sympatric speciation
formation of 2 different species from 1 original species due to reproductive isolation, in which the population inhabit the same geographical location
artificial selection
selective breeding of organisms, which involves humans choosing the desired phenotype of an organism & interbreeding the desired phenotypes, therefore selecting the genotypes that contribute to the gene pool of the next generation
in artificial selection, what is the selecting agent?
humans
inbreeding depression
reduced biological fitness in a given population as a result of inbreeding, or breeding of related individuals
why does artificial selection cause inbreeding depression?
1. at each stage of the selective breeding programme, individuals with the desired traits are selected
2. individuals with undesirable characteristics aren't selected
3. this will decrease genetic diversity in the gene pool
4. and increase the chances of an individual inheriting 2 copies of a recessive allele
hybrid vigour
Increased heterozygosity giving increased vigour of fertility growth & survival
phenotypic traits that have been selected for in dairy cows
long lactation period, high milk quality, large udders, resistance to disease, calm temperament, large yield / volume of milk
modern techniques used in the selective breeding of animals
artificial insemination, IVF, progeny testing, use of a surrogate mother, cloning, use of gene probes
genome
an organism's complete set of DNA, including both the genes & the non-coding sequences of DNA
DNA sequencing
a technique that allows genes to be isolated and read
what are the ways in which DNA can be manipulated?
DNA profiling, genomic sequencing, genetic engineering & gene therapy
the process of gene sequencing
1. genes are mapped to identify which part of the genome they come from
2. samples of the genome are sheared into smaller sections approximately 100 base pairs long
3. samples are placed into separate BACs & transferred to E. coli cells
4. as the cells grow in culture, many clones of the sections are produced. These are clone libraries
5. DNA is extracted from the BACs & replicated by PCR
6. DNA is digested & cut up by restriction enzymes into smaller fragments
7. the fragments are separated into size order by electrophoresis
8. computer programmes are used to reassemble the full BAC sequence by analysing the overlaps shown by the fragment sequences
what was DNA research like in the early 1970s?
- DNA structure was known
- base triplets for aas were known
BUT
- the sequence of triplet bases in a gene was difficult to determine
- early work involved working with mRNA, and not DNA
- RNA is unstable because it's single stranded, sequencing a gene using RNA was very slow & only worked with short genes
- in 1975, Fred Sanger developed a method for sequencing the whole genome
the stages in the Sanger method for sequencing DNA (automated DNA sequencing)
1. add a single stranded DNA sequence, primer, DNA polymerase & the 4 nucleotide bases to 4 separate dishes
2. the primer anneals at the 3' end of the DNA strand, allowing DNA polymerase to bind
3. DNA polymerase adds free nucleotides by complementary base pairing
4. to each dish add a modified DNA nucleotide that is labelled with a radioactive isotope
5. the addition of the modified base causes the enzyme to be thrown off, so semiconservative replication is terminated
6. the process is repeated & many different sized DNA strands are created due to the random joining of modified nucleotides
7. electrophoresis is used to separate the DNA fragments by size
8. short chains travel the furthest
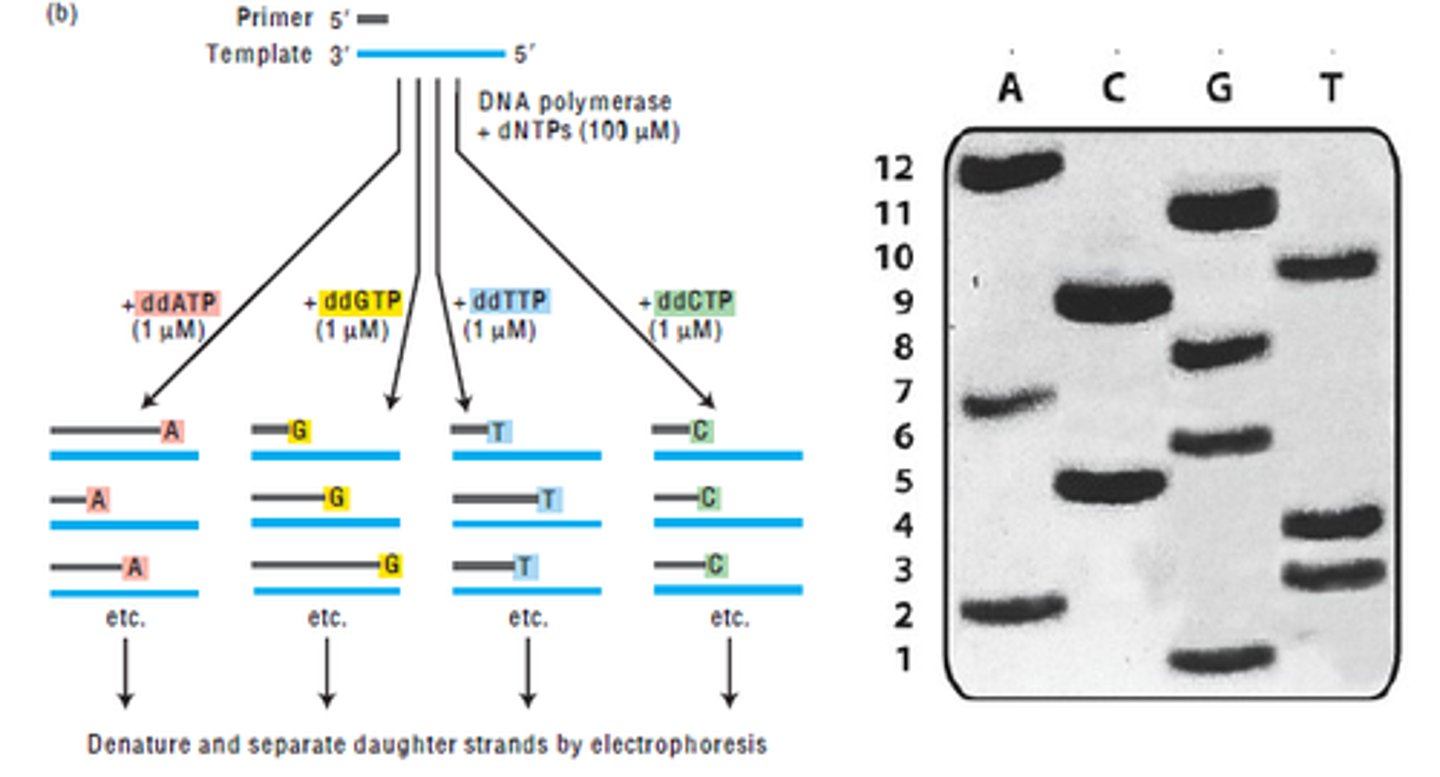
what is the strand produced by the Sanger method?
it is a strand complementary to the initial single stranded DNA sequence
high throughput sequencing
fast, cheap methods to sequence genomes developed in the 21st century
the stages in pyrosequencing (a type of high throughput sequencing)
1. a length of DNA is cut into fragments of 300 - 800 base pairs using a nebuliser
2. fragments are degraded into single stranded DNA.
3. these are the template DNAs & they're immobilised
4. a sequencing primer is added
5. DNA is incubated with DNA polymerase, ATP sulfurylase, luciferase, apyrase, APS & luciferin
6. 1 of the activated nucleotides ATP, TTP, CTP, or GTP is added
7. the nucleotides form a strand complementary to the DNA fragment used as a template
8. this releases 2 phosphorlys as a pyrophosphate
9. APS combines with pyrophosphate to form ATP. This is catalysed by ATP sulfurylase
10. in the presence of ATP, luciferase converts luciferin to oxyluciferin
11. visible light is generated in this reaction and detected by a camera
12. light patterns detected indicate amount of ATP and therefore DNA sequence
applications of DNA sequencing
- comparisons between species
- comparisons between individuals
- predicting aa sequences
- synthetic biology
bioinformatics
The collection, classification, storage, and analysis of biochemical & biological information using computers especially as applied in molecular genetics & genomics
why may bioinformatics be useful in determining whether a newly sequenced allele causes a genetic disease?
- the base sequence of a normal allele & known alternatives are held in a database
- computational analysis allows rapid comparison of sequences with the newly sequenced allele
- aa sequences and protein structures are also held in the database
- computer modelling of the new protein structure can be done from the base sequence
epigenetics
the study of changes in organisms caused by modification of gene expression, rather than alteration of the genetic code itself
what do the 0.1% differences in DNA between humans arise from?
substitutions called Single Nucleotide Polymorphisms
what can often cause a modification of gene expression?
methylation of DNA, which causes transcriptional silencing. Methods to map the methylation of whole human genomes can help researchers understand the development of certain diseases (e.g. cancer)
how is gene sequencing used to predict aa sequences?
- it's easier to sequence an organism's genome than polypeptides
- if researchers know which gene codes for a specific protein, they can use knowledge of which base triplets code for which aas to determine the primary structure of proteins
- the researchers need to know which part of the gene codes for introns & exons
synthetic biology
the area of biology concerned with designing & building useful biological devices & systems. It includes biotechnology, evolutionary biology, molecular biology & biophysics
goals of synthetic biology
- build engineered biological systems that store & process information
- provide food
- maintain human health
- enhance the environment
uses of synthetic biology in information storage
scientists can encode vast amounts of digital information onto a single strand of synthetic DNA
DNA profiling
a forensic technique used to identify individuals by characteristics of their DNA
stages in DNA profiling
1. DNA is obtained from an individual e.g. by a mouthswab
2. restriction enzymes digest the DNA & cut it into fragments at specific recognition sites
3. the fragments are separated by gel electrophoresis & stained to obtain a banding pattern
4. the banding pattern is compared to another individuals', which has been treated with the same restriction enzymes
5. related individuals will have more similar banding patterns
applications of DNA profiling in disease
protein electrophoresis can identify the type of Hb produced by an individual, and can be used to diagnose sickle cell anaemia