(26-28) tRNA+ Protein synthesis
1/46
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
47 Terms
tRNA is the — of the functional RNAs at — to — nucleotides in length
smallest; 75 to 85
tRNA function
to take an individual amino acid to its appropriate location to be attached to an amino acid chain
Amino acids are attached to tRNA by a specific —. These molecules are each specific to one —
aminoacyl synthetases; tRNA family
There are about — different tRNA molecules that fit into — families. There is a family of tRNA for each —
50;20; amino acid
Draw and label a tRNA with an attached amino acid
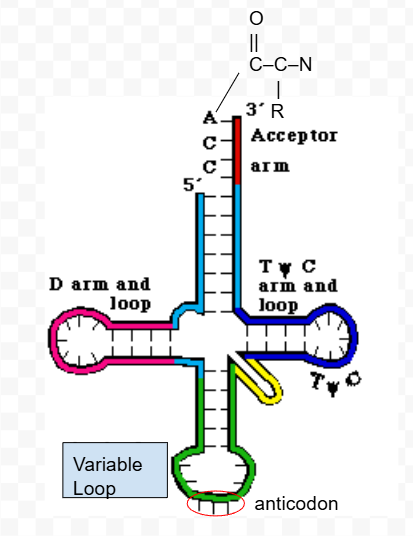
D-loop
hairpin loop on a tRNA closest to the 5’ end. Has modified nucleotides (specifically uracils are more modified).
TψC loop
hairpin loop on a tRNA closest to the 3’ end. Has modified nucleotides (specifically uracils are more modified).
Variable loop
hairpin loop on a tRNA between the D-loop and TψC loop. Contains the anticodon which is important for identifying the tRNA as the correct tRNA for building a specific protein.
Where does an amino acid bind to on a tRNA?
at the 3’ end, to the adenine nucleotide via the carboxyl carbon
What is a tRNA with it correct attached amino acid called?
aminoacyl tRNA
If the wrong amino acid is attached to a tRNA, it is often — to the cell/organism
deleterious
Why is it uncommon for the incorrect amino acid to be added to a tRNA?
there are mechanisms/enzymes to ensure that doesn't happen
At the 1st active site of amino acyl synthetase, what happens? 2nd active site? (show 2 equations)
1st active site- the amino acid is energized via ATP; 2nd active site- the energized aa is bound to tRNA.
The fidelity of aminoacyl synthetases are enhanced by what feature of the enzyme?
it having two active sites
3 steps of protein synthesis
initiation, elongation, termination
IF1
initiation factor 1. With IF2, it assists and binds to the first tRNA holding methionine to bind to the AUG start codon of the mRNA
IF2
initiation factor 2. With IF1, it assists and binds the first tRNA holding methionine to bind to the AUG start codon of the mRNA. May be phosphorylated to become inactive. This allows for regulation of protein synthesis.
IF3
initiation factor 3. Aligns mRNA to bind to 40s ribosomal subunit at the 5’ ribosome binding site at the start of initiation. Splits 60s and 40s ribosomal subunits in termination.
Steps of protein synthesis initiation
ribosome binding site at 5’ end of mRNA binds to 40s ribosomal subunit with the help of IF3. tRNA with methionine and anticodon UAC binds to AUG codon on the mRNA with the assistance of IF1 and IF2. the 60s (large) ribosomal subunit binds and the initiation factors disappear forming the 80s ribosome.
What is always the first codon to be translated? This codes for what aa?
AUG; methionine.
What is the anticodon of the first translated codon?
UAC
If the ribosome binding site is nucleotides 1-50, where is the start codon?
at nucleotides 51-53
t/f you may see an 80s ribosome in initiation
false, the formation of the 80s ribosome indicates the end of initiation.
t/f the 80s ribosome is somewhat hollow, like a wiffle ball
true
The interior of a ribosome may have — nucleotides of mRNA at one time
20
t/f it is ineffective to control protein synthesis at the initiation step
false
What are the “rooms” of the large ribosomal subunit
A site and P site
What site of the large ribosomal subunit does the methionine tRNA start in after initiation?
P-site
A site
acceptor site. Where new tRNAs enter the ribosome and bind via their anticodon to the next codon of the mRNA.
P-site
peptidyl site or donor site. The site at which the aa of tRNAs are added to the chain
Tu
temperature unstable protein. Elongation factor. Along with Ts, escorts tRNAs to A-site.
Ts
temperature stable protein. Elongation factor. Along with Tu, escorts tRNAs to A-site.
Peptidyl transferase
enzyme. Physically associated with 60s ribosomal subunit. Breaks bond between carboxyl carbon and 3’ end of tRNA in the P site to form the peptide bond between the amino acid and the amino acid in the A site
G-translocase
enzyme. Responsible for shifting mRNA one codon at a time through the ribosome as each aa is added to the chain using GTP for energy.
Steps of elongation
tRNA is guided to A site by Tu and Ts. GTP is hydrolyzed to force tRNA into appropriate locations (bound to 5s rRNA of large/60s subunit). Peptidyl transferase breaks the bond between the 3’ end of the tRNA and carboxyl carbon of the P-site tRNA and forms the peptide bond between the P site amino acid and A site amino acid (amino acids are added to the carboxyl end of the P site aa ). GTP is hydrolyzed as G-translocase shifts the mRNA one codon through the ribosome and the tRNA in the P site exits the ribosome and the one in the A site shifts to the P site.
The ____ end of the peptide chain found in the p-site is added to the ___ end of the amino acid found at the A site by the — protein . It is said that the protein is always growing from the ___ end.
Carboxyl terminus, N terminus, peptidyl transferase, Carboxyl terminus.
As the amino acid chain elongates and it sticks out of the ribosome, it — to keep stable.
folds
Draw a aa chain in the process of elongation with 3 amino acids (label the the amino acids in order that they were added to the chain)
3 stop codons
UAG, UAA, UGA
t/f stop codons are recognized by tRNAs
false, they are recognized by releasing factors
R1
releasing factor 1. Recognizes UAG or UAA. causes aa chain and P-site tRNA to detach from the ribosome.
R2
releasing factor 2. Recognizes UAA or UGA. causes aa chain and P-site tRNA to detach from the ribosome.
Steps of termination
stop codon is recognized by a releasing factor which acts as a “bowling ball” and causes the P-site tRNA and protein to detach from the ribosome. IF3 then causes the 60s and 40s ribosomal subunits to detach from one another.
Why is it efficient for IF3 to be involved in both termination and initiation
because it is present in the last step of termination and first step of initiation, it allows for protein synthesis to immediately begin again after completion if needed.
Antibiotics typically target — ribosomes which are present in microbes
70s
t/f Antibiotics are harmless to 80s ribosomes
false, they may harm them at high concentrations
Why are some antibiotics slow acting?
they target 70s ribosomes in bacteria which causes them to slowly die as their protein synthesis comes to a halt