DNA Repair
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
What are the spontaneous DNA mutations that require DNA repair?
Depurination
Deamination
spontaneous oxidative destruction
uncontrolled methylation
hydrolytic attack
UV expose (non-spontaneous I think)
Double stranded DNA breaks
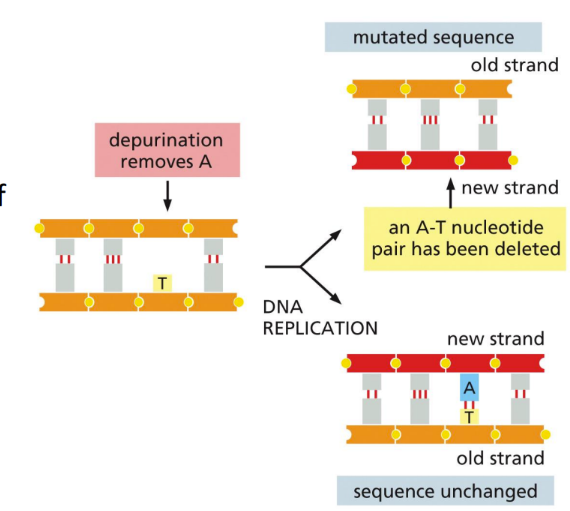
What is depurination?
Spontaneous loss of ~5,000 purines (A or G) bases lost per cell per day because of hydrolysis of the glycosyl linkage to deoxyribose
Occurs when the bond connecting the purine to the sugar is broken by a water molecule
Resulting in a purine free nucleotide that can’t act as a temple during replication
A hydroxyl group is left instead of the nitrogenous base group
MOST COMMON
Can result in a complete loss of both bases (deletion of base pairs)
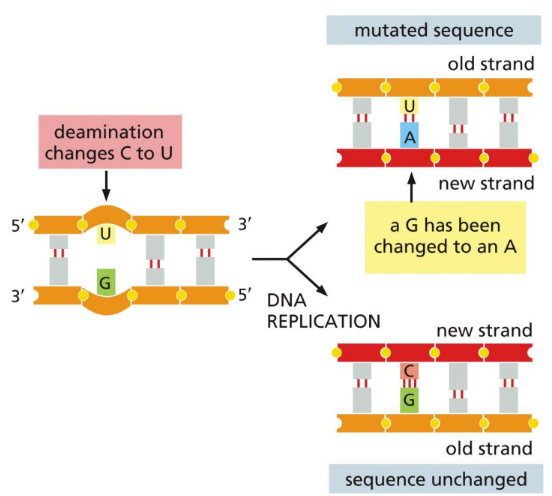
What is deamination?
Spontaneous removal of the amine (NH2) group from ~100 cytosines per cell per day to form uracil
Occurs when hydrolysis wipes away the NH2 from a cytosine and replaces it with an oxygen. Forming an uracil and an ammonium ion
C => U
Changes the base pairing
What is spontaneous oxidative destruction?
Reactive oxygen radicals or chemicals exposure cause base damage
Red arrows
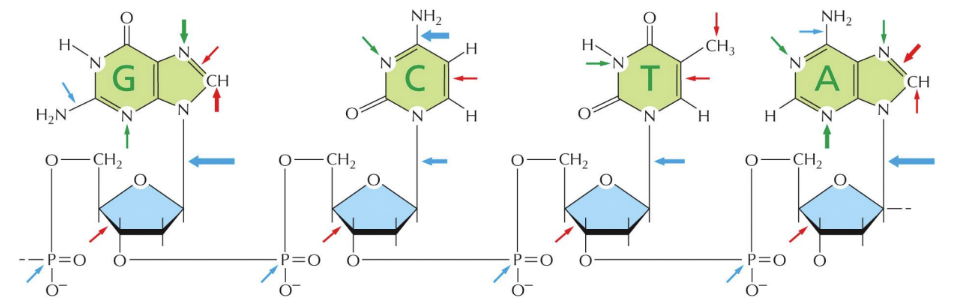
What does the green arrows represent
Uncontrolled methylation
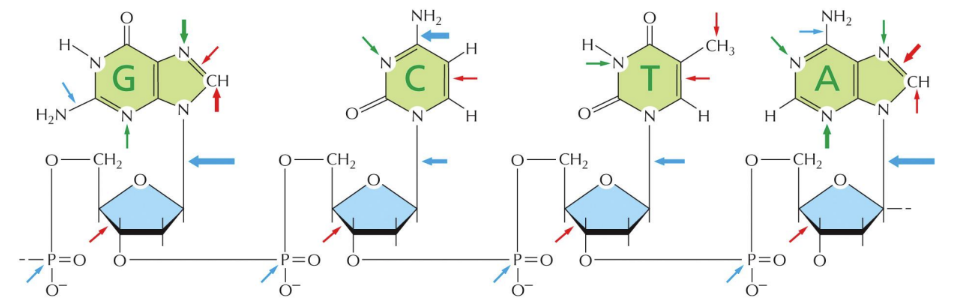
What do the blue arrows represent
Hydrolytic attack
What is UV exposure (non-sponantous)?
Forms pyrimidine dimers via covalent linkage between two bases
A covalent bond is made between 2 bases
Pyrimidine dimers = can block transcription/replication when brought close together and not repaired
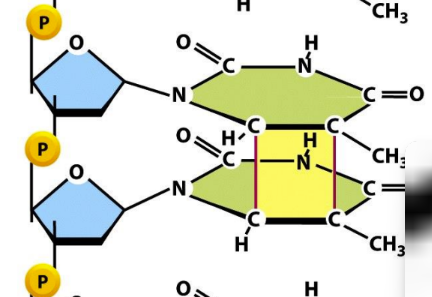
What is double stranded DNA breaks
occurs frequently with ionizing radiation issues, replication errors, & oxidizing agent destruction
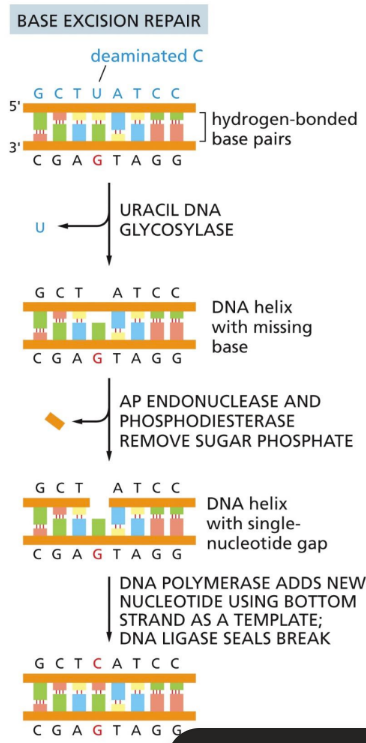
What is BER (Base Excision Repair)?
removed affected base and replacing it with the correct one
DNA glycosylases are specific for altered DNA base (DETECTION)
DNA glycosylase enzyme flips out each nucleotide until it finds one to repair
Steps (REPAIR PROCESS):
Hydrolysis by specific DNA glycosylase
Flips out nucleotide
AP enzymes (aqurinic or apyrimidic endonucleases) chew out sugar backbone here
DNA polymerase adds new nucleotide
DNA ligase seals nick with new sugar piece
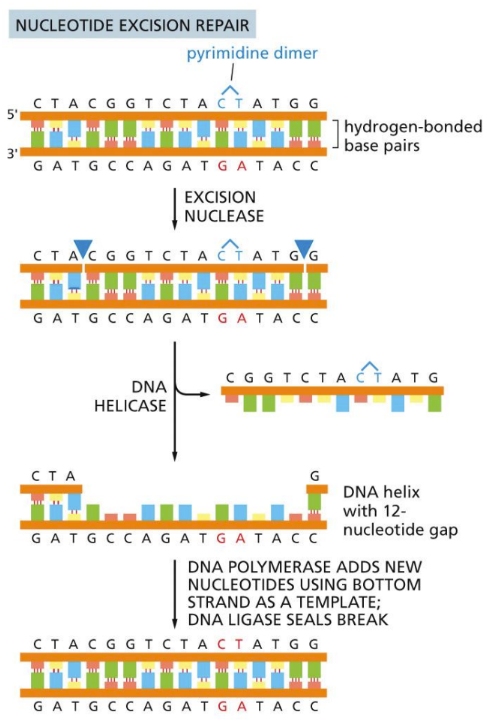
What is NER (Nucleotide Excision Repair)?
Removing a ~30 nucleotide chunk of affected strand
A protein recognize a dimer begins this repair (DETECTION)
The errors produce a atypical “bulky lesion” (not normal for DNA helix
Large multienzyme complexes see the lesion and cleave out the phosphodiester backbone
DNA pol. Helicase, and ligase go to single strand to copy over
Steps (REPAIR PROCESS):
Helicase unwinds local duplex
Excision nuclease cleaves on both sides of damage leaving a gap of ~30 nucleotides
DNA polymerase and ligase finish job
What is transcription-coupled repair?
Errors repaired once transcription has started (past replication)
RNA polymerase will stall transcription where it finds a DNA error
It'll reverses itself and call over coupling proteins
Coupling proteins will repair the error (direct excision repair machinery (calls over NER)) and leave
RNA polymerase continues transcribing
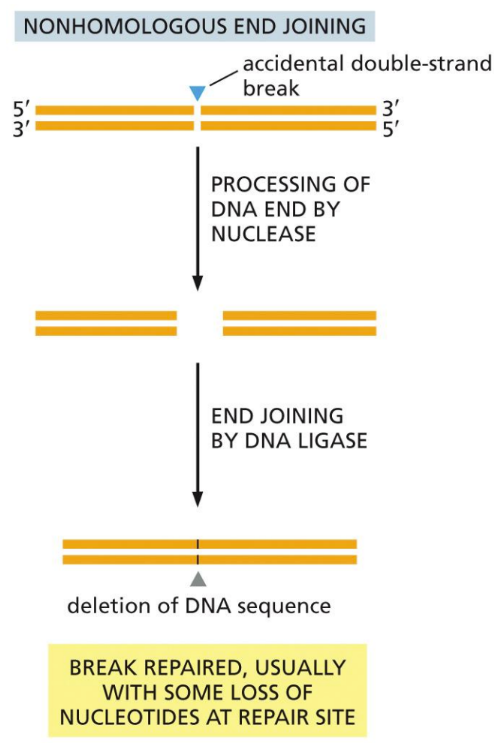
What is NHEJ (Non-homologous end joining)
Repairs double stranded DNA breaks by removing a set of nucleotides, then ligating the strands back together (does delete part of strand)
DNA polymerase µ & λ (family X) involved
Negative side effect = often results in deletion mutations or translocations
Acceptable because majority of genome is non-gene coding
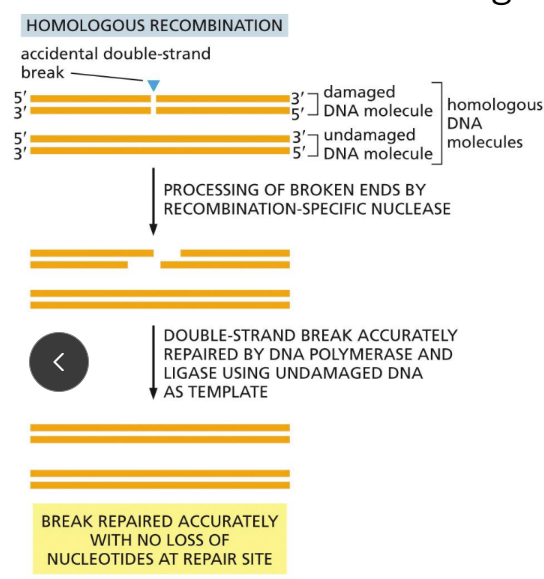
What is HR (homologous recombination)?
repairs double stranded DNA breaks
A more accurate mechanism (than NHEJ)
Occurs only AFTER replication (S and G2 phase) has occurred, when there is a nearby duplex daughter DNA to serve as the template
Negative side effects =
Loss of heterozygosity
Possession of these proteins is often controlled by other proteins like BRAC1/2
These alterations can cause cancers
How does DNA base chemistry facilitates DNA damage detection?
What is XP(Xeroderma Pigmentosum)? phenotype, process affected, and mutation
Thymine dimers (made via UV damage) block transcription and replication because the repair enzymes are mutated
Dysfunctional repair enzyme that is HERITABLE (germ line mutation)
Can result in tumors
Phenotype: Skin cancer, UV sensitivity, and neurological abnormalities
Non-ionizing radiation forms the dimers and there isn’t a repair enzyme available to fix them
Process affected: NER (dysfunctional repair enzyme)
What is Cockayne Syndrome? phenotype, process affected, and mutation
Defect in transcription-coupled repair
Mutation in 2 genes on chromosomes 5 & 10
Phenotype: UV sensitivity, developmental abnormalities (underdeveloped tissues/organs)
Process affected: coupling of NER to transcription
Because there is so many lesion sites (errors) RNA polymerase gets stalled and there is insufficient genes (certain genes)
Lesions are from UV damage
What is AT? phenotype, process affected, and mutation
Single nucleotide base change (point mutation (via deletion, insertion, or substitution))
Phenotype: Leukemia, lymphoma, genome instability, and gamma-ray sensitivity
Process affected: ATM protein (a protein kinase activated by double-strand DNA breaks
What is BRCA1? phenotype, process affected, and mutation
Spontaneous mutation on brca1 gene
Phenotype: Breast and ovarian cancer
Process affected: Repair by homologous recombination
What is the consequence of a loss of DNA repair gene
Unrepaired spontaneous errors can become permanent if they don’t get fixed
DNA repairs depends on healthy DNA repair genes to make functional repair enzymes/proteins
Many repairs in 24hrs (more than 1,000 for some)
Semi-conservative replication/replication allows the “good” strand can be used to restore damage or continue functioning as normal
What are the DNA polymerases involved in DNA repair?
Family B (α, ε, & δ) = Replication
Family X (β, µ, & λ) = BER/NER repair
Family Y (n, l, & k) = translesion polymerases
Don't have exonucleolytic activity
Don't discriminate about which nucleotide to add
Often leads to errors
Come and rescue Family B polymerases when they are stuck trying to replicate
Once pushes through error, will leave strand so DNA polymerase family B can continue