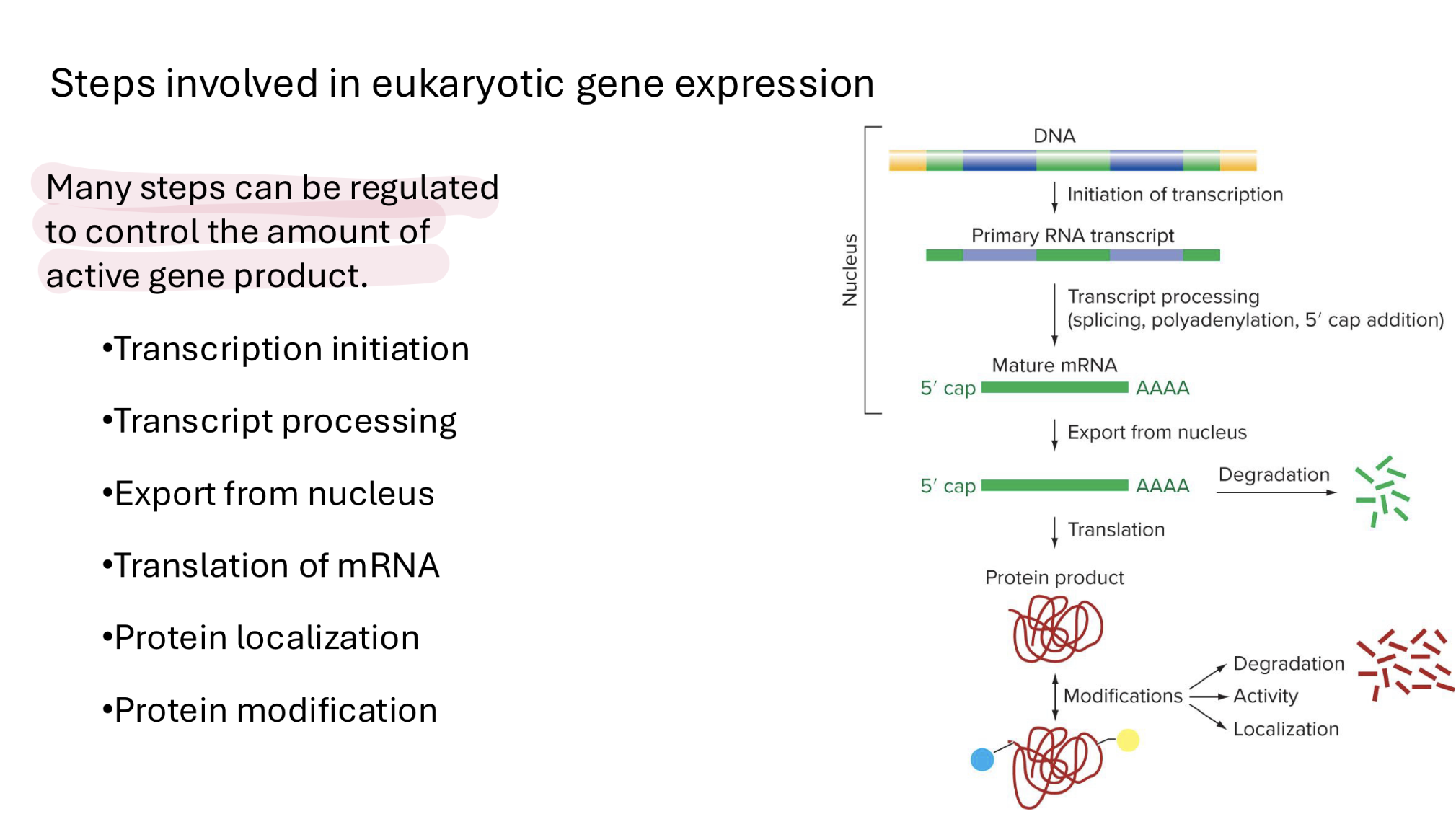
Eukaryotic Gene Expression (Midterm 3 Lesson 4)
1/71
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
72 Terms
t/f: eukaryotic gene expression is more complex than prokaryotic
true
why is eukaryotic gene expression more complex than prokaryotic
multiple organs
lots of diff organs/tissues that need certain things to be active
have transcription in nucleus then translation in cytoplasm
have chromatin (w DNA wrapped around histones)
t/f: eukaryotes organize their genes into operons. explain why or why not
false
what specifically about the transcription process in eukaryotes makes it more complicated than in prokaryotes
their control of transcription
many steps can be regulated to control the amount of active gene product in eukaryotes as opposed to prokaryotes
how many diff RNA polymerases transcribe diff types of genes in eukaryotes
3-5
what are protein-encoding (structural) genes transcribed by
RNA polymerase II
what is a DNA sequence upstream of a gene that binds proteins that modulate the transcription of that gene called
a cis-acting factor
factor that modulates the regulates a gene that is on the SAME DNA MOLECULE AS IT
dna sequences in general are usually cis-acting factors while proteins are trans-acting
dna sequences can’t migrate to other DNA sequences but proteins can
what do trans-acting proteins bind to
cis acting regulatory regions
what are transcription factors
sequence-specific DNA binding proteins
bind to promoters and/or enhancers
recruit other proteins to influence transcription
what are the 3 types of transcription factors
basal factors
activators
repressors
what are the major cis-acting regulatory elements
promoters
enhancers
where are promoters located, what are they composed of, what do they bind to, and what can they do on their own
close to gene’s coding region (around 30bps upstream of start site), its a conserved sequence of TATA boxes (sections high in As and Ts)
binds RNA polymerase and basal factors
maintain a basal level of transcription on its own
what are basal factors, what are some examples, and what do they do? what is their mechanism of action
they maintain a basal level of transcription when binding to promoters of protein-coding genes
proteins that are essential for the initiation of transcription in eukaryotic cells.
called "basal" because they are required for basic transcription to occur—i.e., they are needed for RNA polymerase II to begin transcribing most genes, even without any additional regulatory proteins
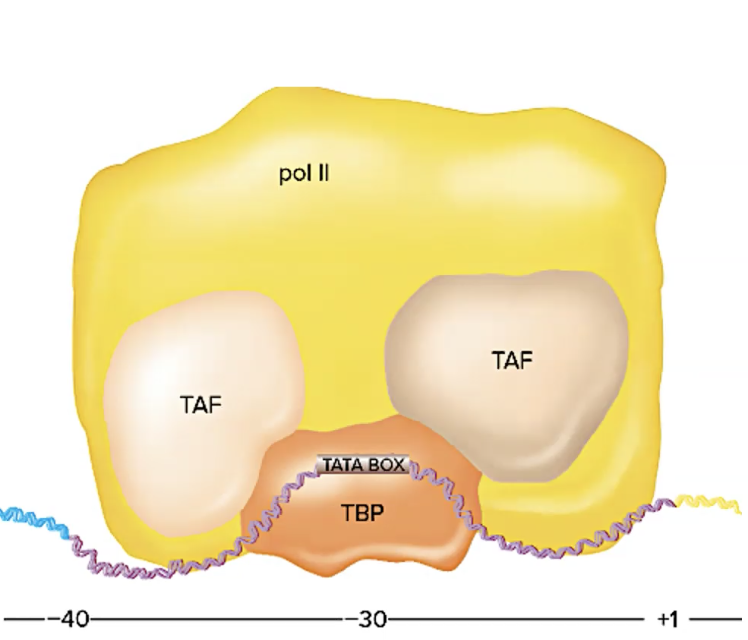
explain ordered binding at the promotor of eukaryotic cells
eg the TFIID complex → TFIID is one of the general (basal) transcription factors required for transcription by RNA polymerase II
ordered binding at the promoter (occurs in this order)
TATA box-binding protein (TBP) binds to TATA box (TBP = one basal factor)
TCP-associated factors (TAFs) bind to TBP (TAF = basal factor)
RNA pol 2 binds to TAFs
TFIID = TBP + multiple TAFs
TFIID is a basal factor
note: is only rna pol 2 and basal factors are bound to promoter, the gene maintains a “basal level of transcription” when this happens
what is a basal level of expression and what type of studies show this
the minimal level of transcription that occurs in the absence of activators and repressors
in vivo studies show basal levels of rna
what do in vitro/in vivo studies show about basal levels of transcription (no activators/repressors)? explain
in vitro (in test tube) → shows basal levels for most eukaryotic genes is high
in vivo (in acc organism) → shows basal levels for most eukaryotic genes is much lower
bc of histones in cells → chromatin has DNA molecules wrapped around histones to form nucleosomes → makes it harder to transcribe
in vitro studies usually j have raw dna in it so it can go faster
what causes the basal level of expression in the absence of both activators and repressors
dna of eukaryotes is packaged into chromatin by the wrapping of dna around histone proteins to form units of nucleosomes → makes it harder to transcribe dna when it is wound like that (proteins for transcription have a really hard time binding bc dna is so tightlyt wound around histones)
NOTE: BASAL FACTORS are things that help rna pol bind to promoter
what are proteins that bind directly to the promoter and are necessary for rna pol 2 binding and transcription initiation?
basal factors (eg tata binding protein)
what are some methods gene expression is controlled by
repressors and activators
repressors can recruit co-repressors (or not) and activators can recruit co-activators (or not)
chromatin structure
acetylation of histones (turns transcription on)
methylation of histones (can turn transcription off or on)
depends on which AAs, # of methyl groups
dna methylation of promoter (turns transcription off)
what are some diffs in how transcription is “turned off” in prokaryotes and eukaryotes
Bacteria:
transcription is actively turned off by repressor proteins binding to the DNA
Eukaryotes:
transcription is passively minimized due to packaging structure of chromatin
passively lowered bc wound so tightly around
transcription rates depend on remodelling of chromatin structure and subsequent interactions w transcription factors
note: still have repressors in eukaryotes but transcription is passively minimized bc of histones ; repressors can LATER provide more specificity but are not main method of action
what are some other things besides rna pol that bind to promoters in prokaryotes and eukaryotes
prokaryotes:
sigma factor (binds and guides rna pol)
sigma factors are a type of basal factor
eukaryotes:
more/diff proteins
TAF
TBP
other proteins that bind to promoter and bind to rna pol
in both cases there are basal factors that bind to promotor in addition to rna pol
what is the basal factor for prokaryotes
sigma factor
what is the basal factor in eukaryotes
TBP TAF
what is the second main cis-acting regulatory element in eukaryotes
enhancers
where are enhancers located
can be really close or really far away from the core promoter (even > 10,000bp away)
can be upstream or downstream of a gene (either5’ or 3’ to the transcription start site)
still functional when moved to diff positions relative to promoter
even if you move it it will STILL HAVE THE SAME FUNCTION
mostly bc dna can loop around itself and make it so enhancer and promoter are close tg regardless of where on the dna the enhancer is located
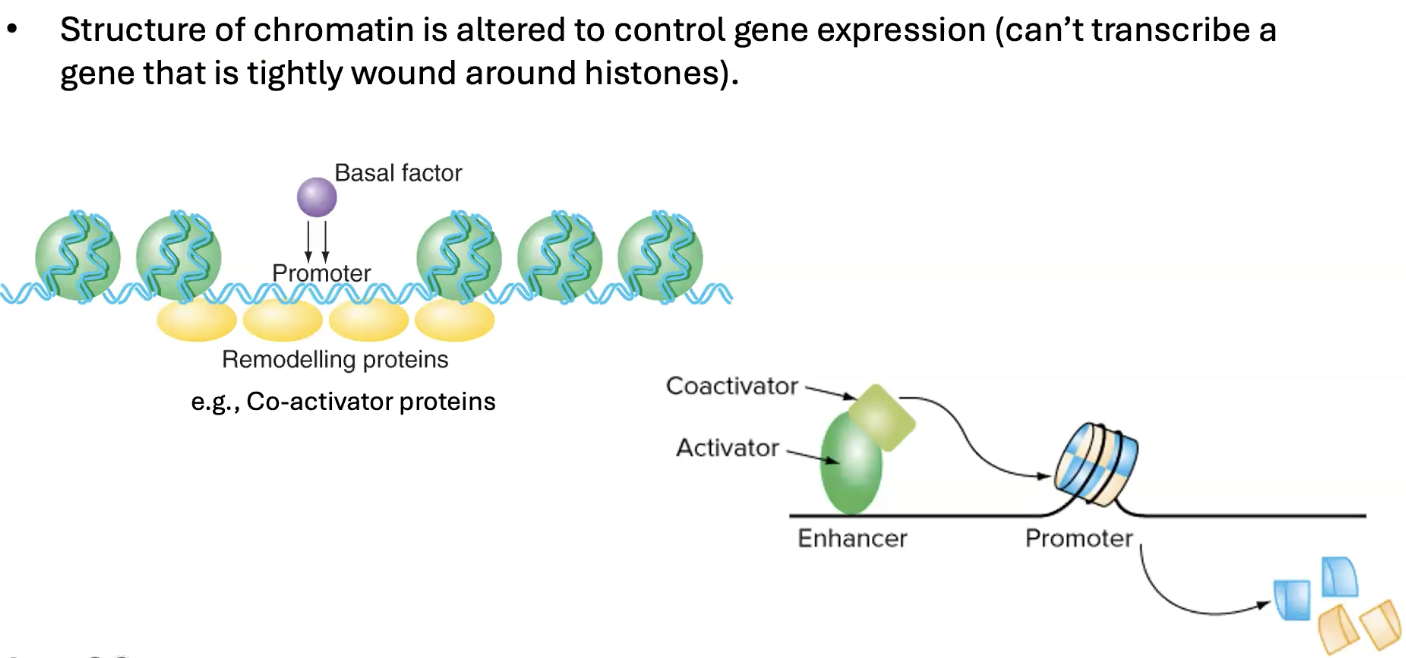
what is the purpose of enhancers
bind transcription factors (activators specifically) that inc the rate of transcription of a gene
pull in transcription factors close to the promotor region so they bind more
what are the 2 domains activators have? what do they do
DNA binding domain (binds to DNA)
transcription activation domains (bind to basal factors or co-activators)
possible third domain → dimerization domain
what are activators
transcriptional activators that bind to specific enhancers and speed up the rate of transcription
stabilize things bound to promoter so their action can be sped up
note: can also bind to repressors and inhibit them depending on the type of activator
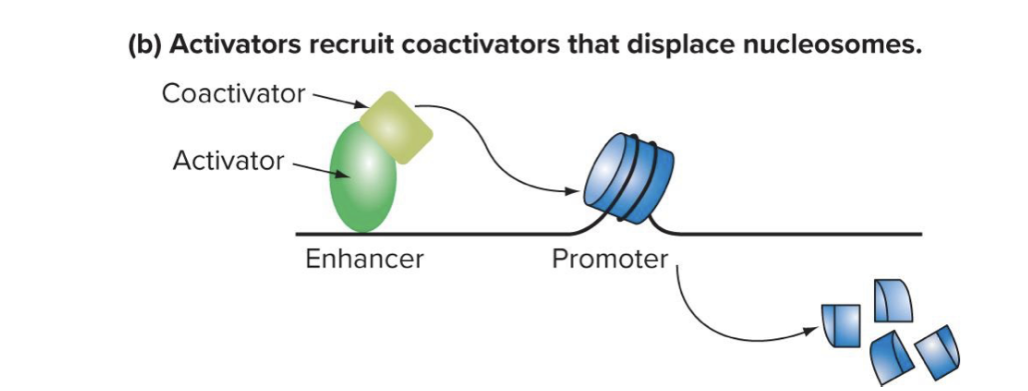
what are the 2 ways activators can work in euks
can recruit co-activators to unwrap DNA of promoter region for transcription
coactivators displace nucleosomes → they clear out the histones and free up the dna
allows basal factors and rna pol to bind
bc eukaryotic dna is wrapped around histones a lot of their regulation has to do w modifying the histones
can recruit basal factors and pol 2 and stabilize the interaction
dna loops around so enhancer and promoter are across from each other
note: is on enhancer for both mechanisms of action
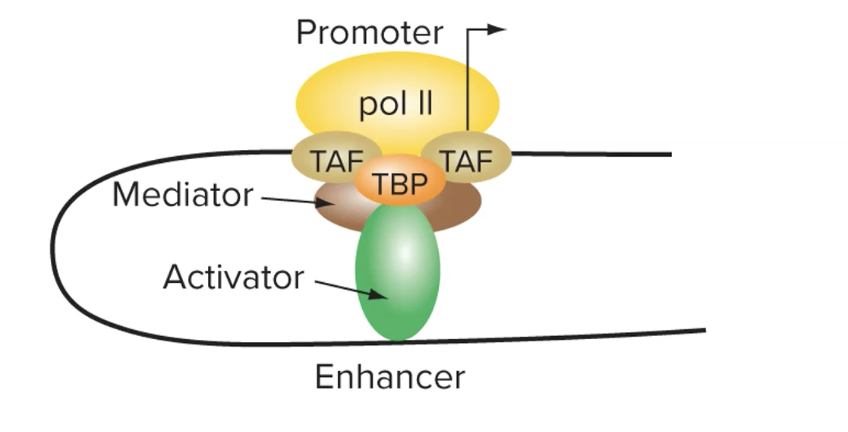
do basal factors bind to both the promotor and enhancer
promotor → yes → needs basal factors to bind to directly allow rna pol to bind
enhancer → no → gets bound to by activators or sometimes repressors
what are these all examples of
basal factors
activators
repressors
different types of transcription factors
what do mediator proteins do
facilitate interaction between the activator and other proteins that are bound to the promotor
what are repressors
proteins that bind to selected sets of genes at sites known as silencers and thus slow transcription
where do activators act
right on proteins that are bound to the promoter (doing so inc rate of transcription by stabilizing their connection to promoter)
able to so so bc dna folds to bring enhancer and promoter close tg
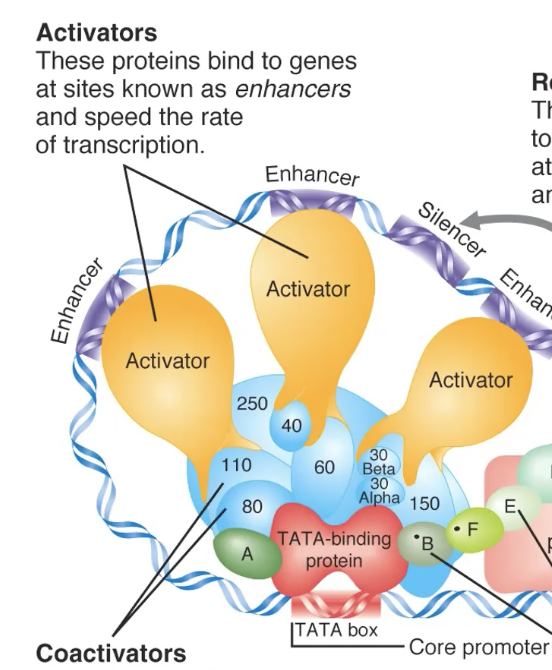
how do distant enhancers interact w genes?
distant regulatory elements can be brought closer tg by chromatin looping
base of the loops held tg by proteins
CTCF
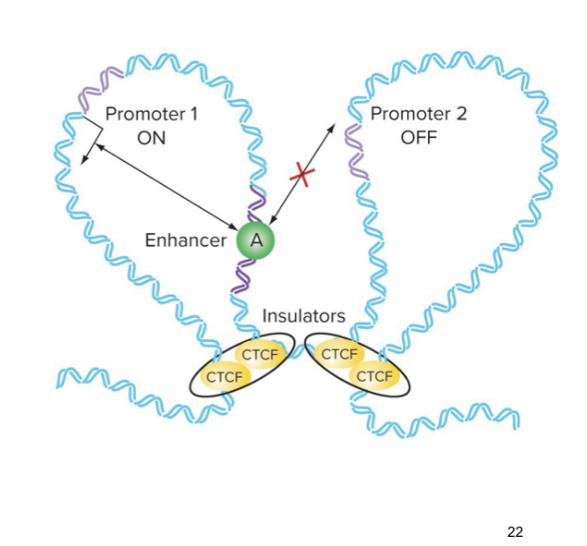
what is CTCF
a zinc finger protein → holds loops of dna tg that bring enhancer and promotor closer tg
what prevents an enhancer from interacting w the wrong promotor
an insulator
what are insulators and what do they do
regions of dna that bind proteins (eg CTCF) that facilitate dna looping
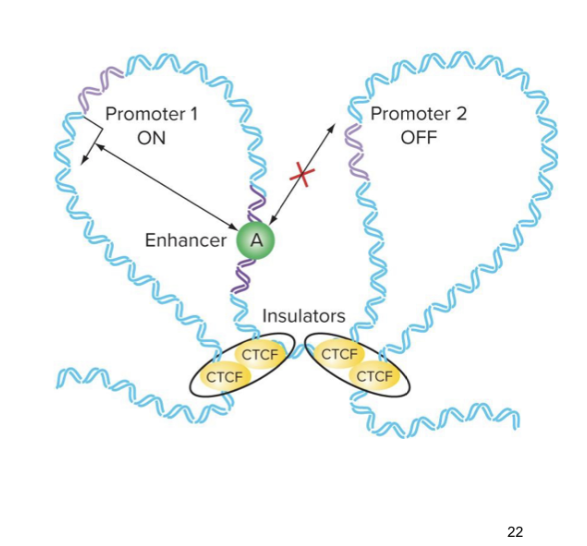
t/f: enhancers for promotors have to be on the same loop
true → it is cis-acting
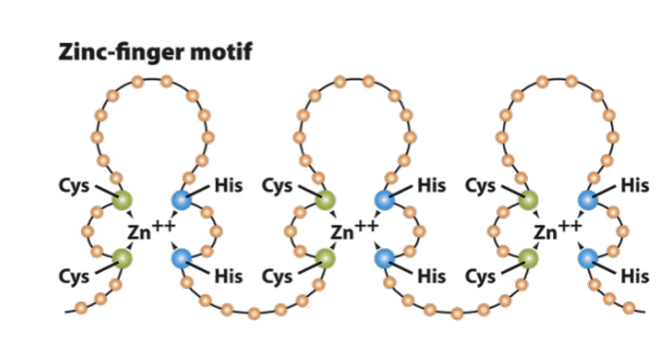
explain what common motifs are in regards to transcription factors
recurring structural patterns (or domains) found in transcription factors that allow them to bind specific DNA sequences and/or interact with other proteins
there are common motifs that are located on diff transcription factors (proteins that regulate gene expression)
eg zink finger
each “finger” on these motifs have a helix that fits int he major groove of dna
the AAs adjacent to the dna determine which dna sequences are recognized
specific zink finger will recognize a specific dna sequence
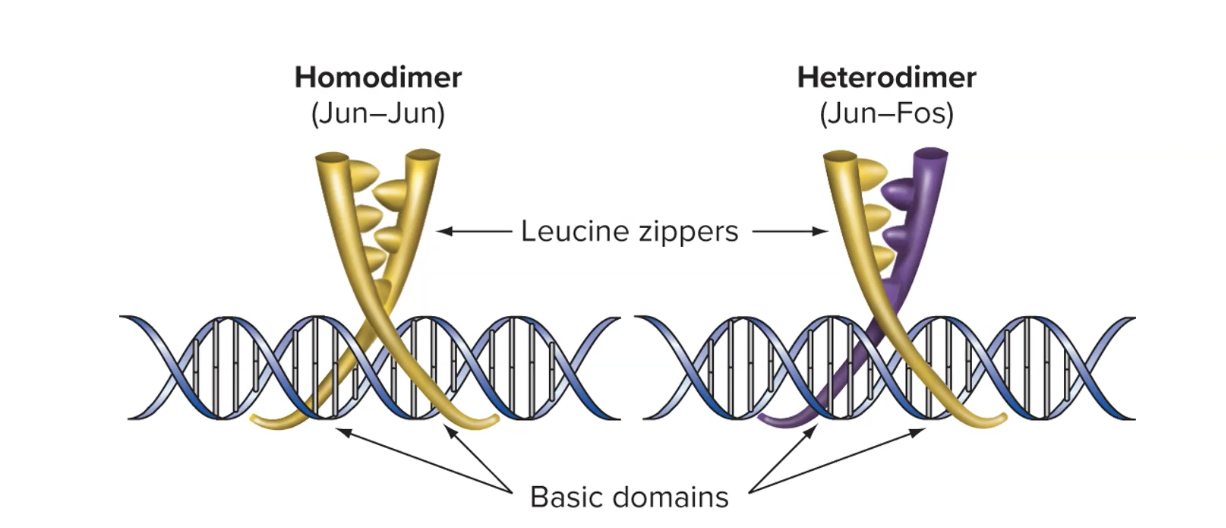
explain the dimerization domain on some transcription factors
many eukaryotic activators need to form dimers (2 molecules bound tg) to fx
happens via the dimerization domain
what is a common example of a dimerization domain on transcription factors
dimers can come tg to form a leucine zipper
each molecule in dimer has leucine residues that bind tg w lucine residues on the other protein
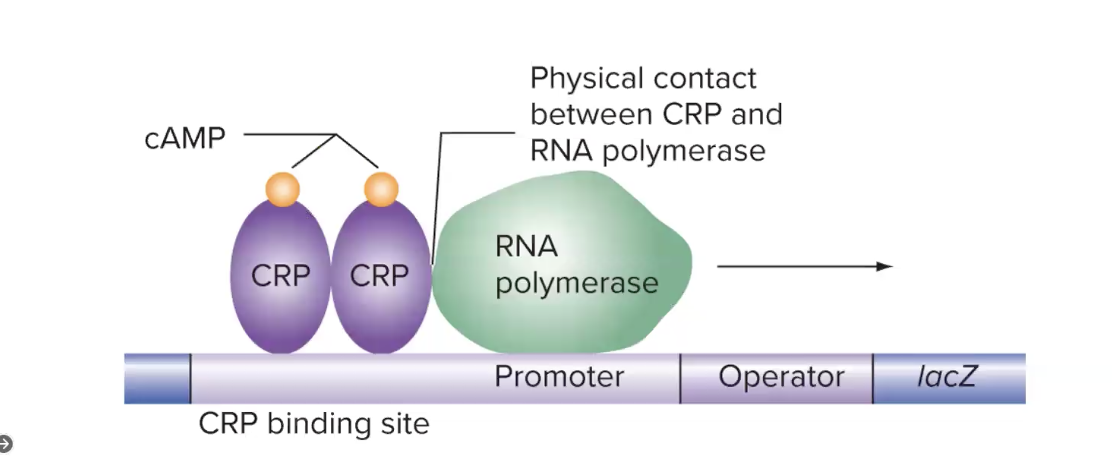
what is an activator that works in the lac operon in prokaryotes
CRP (a positive regulator) of lac transcription
is active when bound to cAMP, its inducer (instead of a coactivator in eukaryotes)
binds to dna and inc efficiency of rna pol binding and activitiy
what are the equivalents to coactivators and corepressors in prokaryotes
prokaryotes have inducers instead
what are some diff ways coactivators can act
bind allosterically to activators and change their conformation
changing histone proteins
interact w proteins at promoter
t/f: coactivators are all proteins
false → estrogens, testosterone, cortisol are all co-activators
explain how some steroid hormones can work as co-activators
testosterone, estrogen and cortisol can all bind to specific transcription factors (activators specifically) → activators in this case are also considered “hormone receptors”
change the receptor (activator’s) shape (allosteric control)
allows them to bind to another molecule and form a dimer
allows them to bind to an enhancer
enhancer loops towards promotor region
inc transcription rate of target gene
note: j one example of what an activator could do (many diff examples)
which molecules are present during basal levels of transcription
basal factors (TBP and TAF)
rna pol
all bound to promoter
nothing bound to enhancer
what do transcriptional repressors do
lower transcriptional activity
what can repressors bind to
enhancers (specific DNA region) ; competition or recruitment of co-repressors)
silencers (specific dna regions)
activator (protein ; quenching)
what are the 2 kinds of repressors
direct and indirect repressors
what do direct repressors do
recruit corepressors that either…
directly prevent rna pol 2 complex (rna pol 2 + TAF + TBP) from binding to promoter
close chromatin (around histones)
what are the 2 ways indirect repressors act
competition
quenching
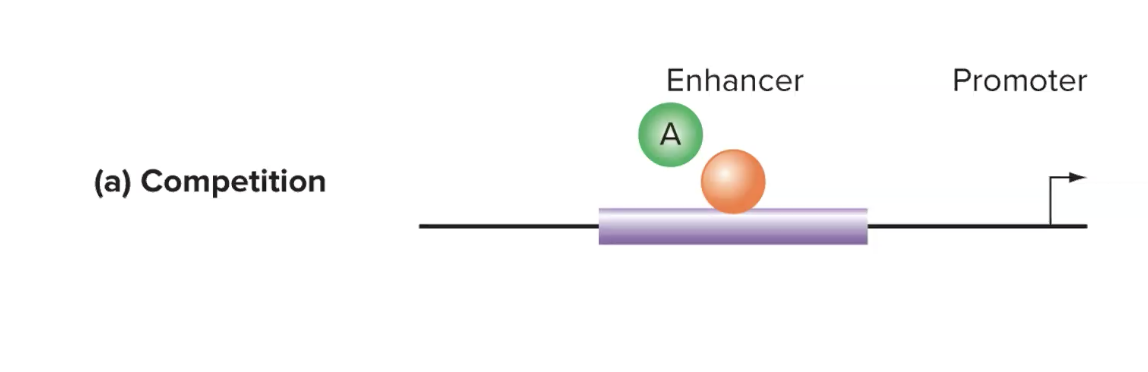
explain how indirect repressors that act through competition work
repressor binds to enhancer and competes w activator
if repressor binds to enhancer, activator can not
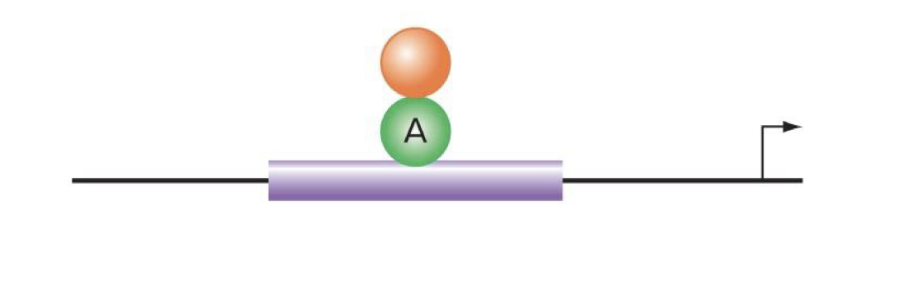
explain how direct repressors that act through quenching work
repressor binds to activator and prevents it from functioning (does NOT bind directly to dna like indirect repressors that act through competition)
what do direct repressors do
actively make sure histones are there and dna is tightly bound
stoping proteins from being able to bind from promoter
what are 2 ways direct repressors can work
recruit corepressors that directly prevent rna pol 2 complex (basal factors specifically) from binding to promoter
can to bind basal complex and prevent them from binding to promoter bc they’re binding to corepressor instead
recruit corepressors that close chromatin
induce wrapping of promoter dna around histones → directly inhibits transcription initiation
t/f: repressors and enhancer can both bind to enhancer and silencer regions of dna
false → both can bind to enhancers, only repressors can bind to silencers
what is chromatin remodelling
when the structure of chromatin is altered to control gene expression (cant transcribe gene that is tightly wound around histones)
can either make easier or harder
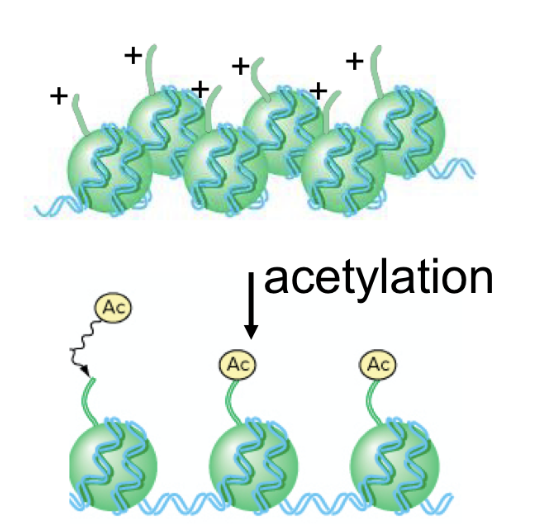
what is histone acetylation
type of chromosome remodelling
acetyl groups get added to histones directly
normal histones have naturally + charges
acetylation adds acetyl groups that have - charges
dna also has - charged groups (ie phosphate groups)
makes it easier to unwind dna and for proteins to bind to promoter
inc rate of transcription
what does histone acetyl transferase do
catalyzes rxn where acetyl group is added to histones → type of coactivators
inc rate of transcription
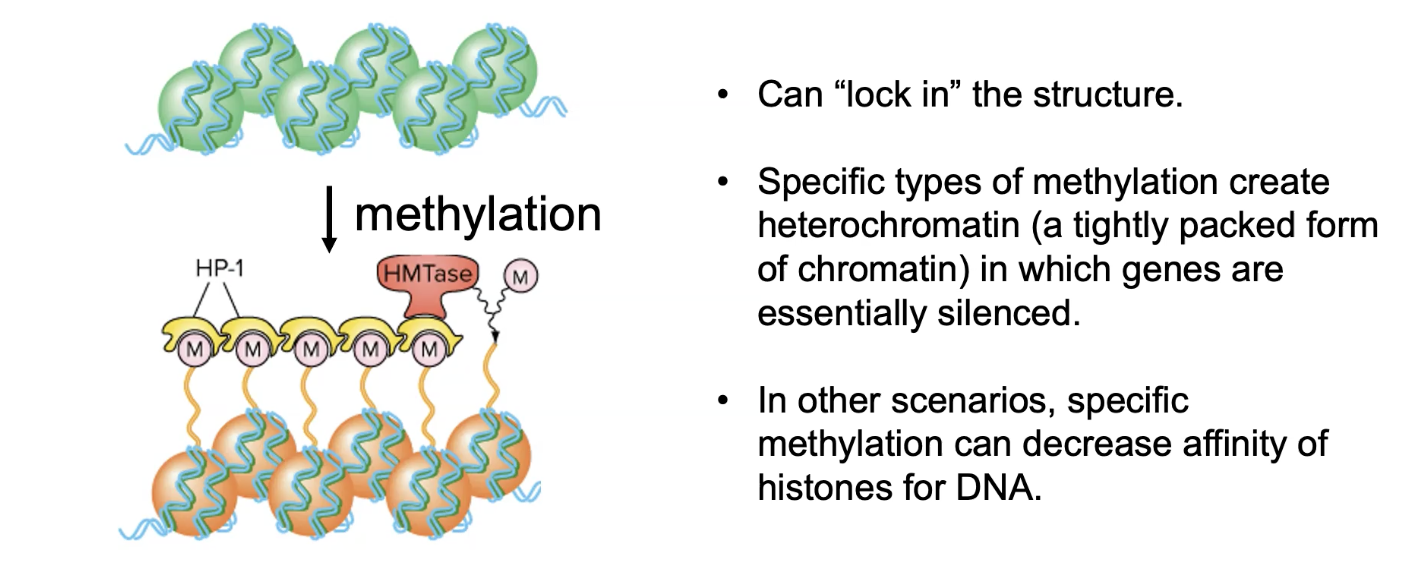
what is histone methylation? explain it and provide a specific example
type of chromosome remodelling
methyl groups are added to histones
has diff influences depending on where its added
mostly results in histones being held closer tg than they normally would be
dec rate of transcription
can be relatively permanent
eg can create heterochromatin (a tightly packed form of chromatin)
in which genes are essentially silenced
basically j permanently turns them off
note: specific methylation can also decrease affinity of histones for dna (co-activators would add in this case)
what does histone methyltransferase (HMT) do
catalyzes addition of methyl groups to histones
is a co-repressor (dec rate of transcription)
what is heterochromatin
tightly-packed form of chromatin (in which genes are essentially silenced)
created by specific types of methylation
which structure on chromosomes contain large regions of heterochromatin
barr bodies → in inactivated x chroms are entirely made of heterochromatin
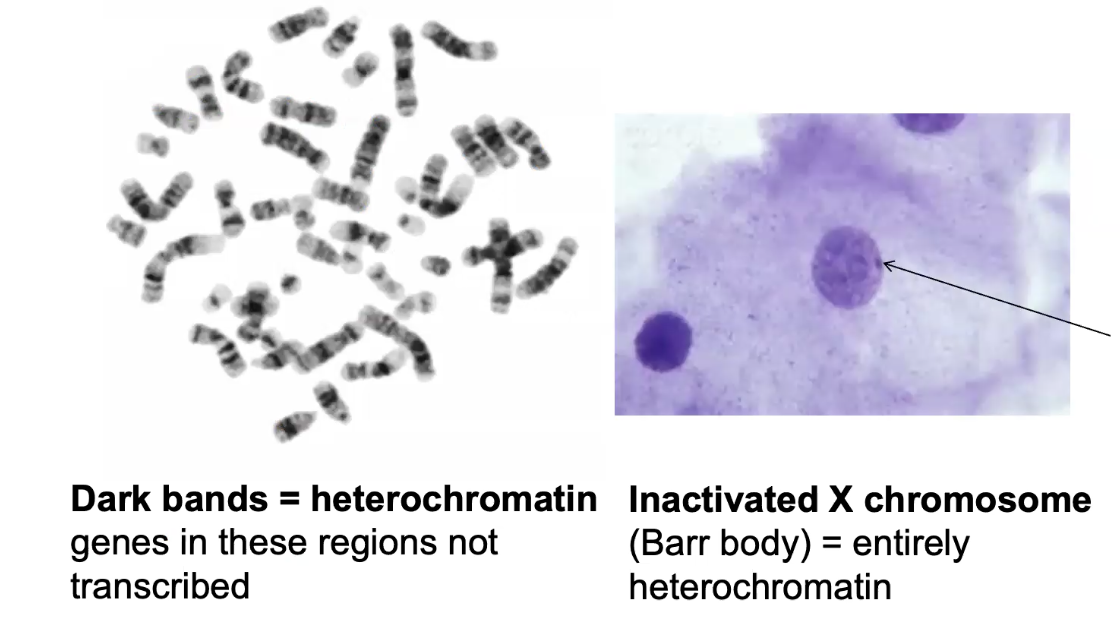
what do dark bands on chromosomes usually indicate
regions with heterochromatin (even more tightly-packed dna) → dna in these regions is not transcribed
what are dna methylation patterns
another way to silence genes
many promoters have blocks of CG-rich sequences called CpG islands (p stands for phosphodiester bonds) → can be methylated and turned off
the acc promoter in the dna sequence itself is methylated → turns it off
what are epigenetics
changes in gene expression NOT related to sequence changes in the dna
eg if you have identical genomes, if both have diff modifications to them (even if they don’t change the sequence itself), they could still have diff phenotypes
eg through diff methylation patterns
if something is cloned, it will not look the same
what are heritable epigenetic markers
epigenetic markers that can be passed down to offspring
what is gene imprinting
specific epigenetic marks (like dna methylation) are placed on certain genes during gamete formation, depending on whether the gamete is sperm or egg → happens naturally, don’t know exactly why → most likely just gene regulation
maintained in mitosis so all cells in body have only one allele expressed
other is silenced
t/f: diet can also influence methylation patterns
true → and they can be inherited
end up w diff phenotypes despite having identical alleles
explain how diff mice w same genotype (heterozygous for coat colour → results in yellow coat) can have different coat colours with the exact same alleles. what experiment was used to text this
remember: yellow gene is lethal when homozygous
2 regions on yellow gene
P1 region: normal gene promoter, weak
methylated = only P2 is expressed, very yellow
P2: second, very strong promoter, can be methylated
methylated = only P1 is expressed, barely yellow
fed 2 pops of hetero mice (yellow) → one w methylated diet, other with regular…
methylated diet (P2 off) were brown, slim, low risk of cancer and diabetes
no methylation (P2 on) were yellow, obese, high risk of cancer and diabetes)