Module 10: Central Dogma I, mRNA production and tRNA
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
What do DNA replication, DNA transcription, mRNA translation all have in common?
Template-based: Use a guide to build (RNA doesn’t need a primer).
Phases: Start slow, build fast, stop at the right spot.
Control: Regulate how often and where to start and stop.
Proteins: Act as switches and volume controls
Explain the RNA polymerase structure with components and function
Structure 1 = Core enzyme:
component: α2ββ' –
Function: Makes RNA from NTPs
Structure 2 = Holoenzyme:
Component: Core + σ – subunit
Function: Finds and binds to the promoter on DNA
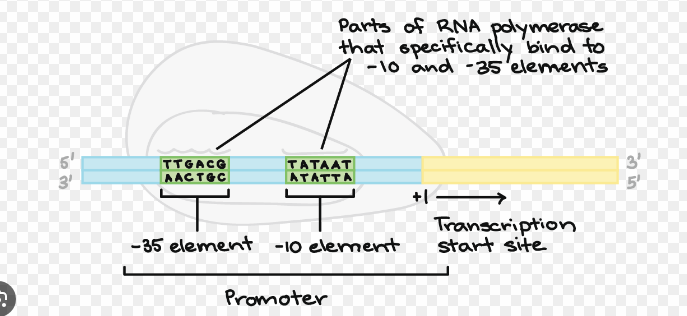
Explain the promoter consensus sequence
The promoter consensus sequence is a short sequence of DNA found in the promoter regions of genes that is recognized by transcription machinery, such as RNA polymerase binding and transcription factors. These sequences are crucial for the accurate initiation of transcription.
These sequences are named based on their position relative to the transcription start site (where the transcription begins). The numbers (e.g., -10, -35) indicate how many base pairs upstream (before) the transcription start site the sequence occurs.
What are the steps in RNA polymerization?
Template binding, open complex formation, chain initiation → elongation → termination
What is constitutive expression?
The continuous production of proteins.
In bacteria, high levels of proteins are toxic
How do bacteria regulate the production of proteins?
by turning gene expression on and off
What is the lactose operon?
The lac operon is a group of genes in bacteria that controls the breakdown of lactose (a sugar). Here's how it works in simple terms:
Lactose absent: When there's no lactose, a protein called the repressor binds to the DNA (the operon) and blocks the genes that would break down lactose. This means the bacteria don’t waste energy making enzymes they don't need.
Lactose present: When lactose is available, it binds to the repressor protein, causing it to change shape and release the DNA. This allows the genes to turn on and produce enzymes that break down lactose for energy.
In short, the lac operon acts like a switch: it turns on when lactose is around and turns off when lactose is absent. This helps bacteria efficiently manage their resources.
What is an operon?
A collection of genes controlled by a promoter and are transcribed as a single mRNA
What are the Lactose operon genes?
lacZ: This gene encodes β-galactosidase, an enzyme that breaks lactose down into glucose and galactose (which the bacteria can use for energy).
lacY: This gene encodes lactose permease, a protein that helps bring lactose into the bacterial cell by transporting it across the cell membrane.
lacA: This gene encodes thiogalactoside transacetylase, an enzyme with a less well-defined role but is thought to help detoxify byproducts from lactose digestion.
What is the Lac repressor protein?
When lactose is present = it binds to the operator region of the lac operon and inhibits transcription of lacZ, lacY, lacA
What is the importance of allosteric changes in the lac repressor?
Allosteric changes in the lac repressor control transcription
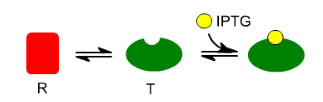
What allosteric changes are happening in the lac repressor?
The lac repressor exists in two states. Relaxed form (red) that bonds to the lac operator. The relaxed form is in equilibrium with the tense form (green).
The tense form has a binding sites for IPTG; when IPTG is present the tense form is favored
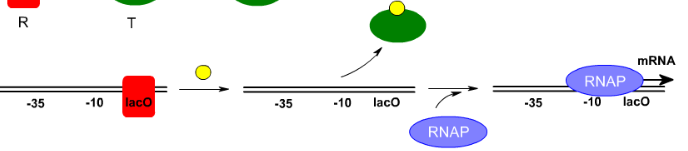
What allosteric changes are happening in the lac repressor?
In the absence of IPTG the lac repressor binds to the lac operator and prevents RNA polymerase (RNAP) from binding
When IPTG (yellow) is present it binds to the relaxed lac repressor (red) and it turns into the tense form (green) and leaves the lac operator and the DNA
The absence of the lac repressor allows RNA polymerase to bind to the DNA and start the synthesis of mRNA
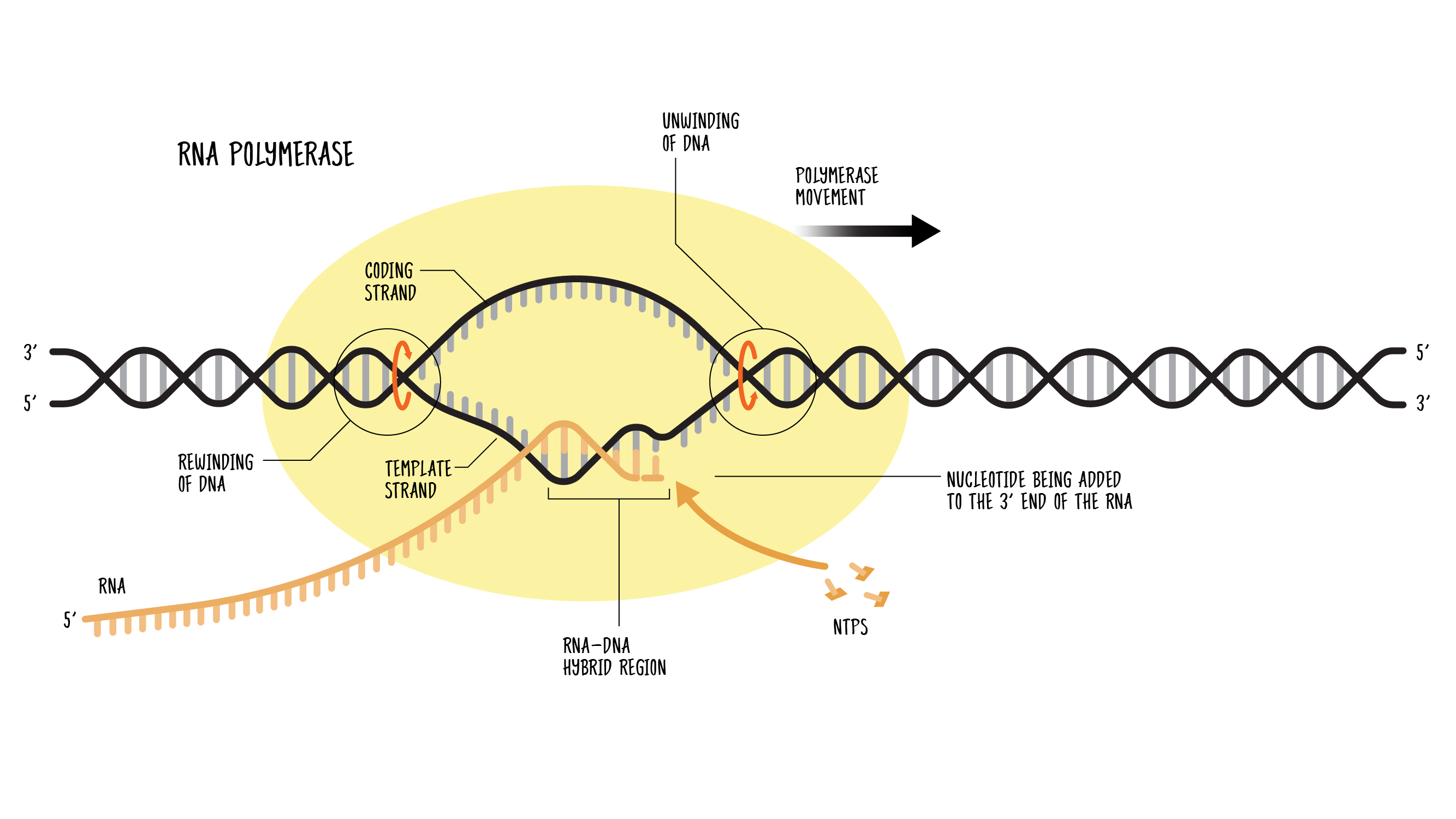
What are the steps of RNA synthesis?
Binding of RNA polymerase
RNA polymerase binds to the promoter region of the DNA, and the DNA strands unwind to form the open complex
Initiation
DNA strand separation (the complex opens) at -12 to +2 bp. The open complex starts the transcription process and initiation begins when 2 rNTPs are formed.
Elongation
RNA polymerase moves along the DNA template strand, adding complementary RNA nucleotides to the growing RNA strand in the 5' to 3' direction. RNA nucleotides are added one by one to the growing RNA strand as the RNA polymerase reads the DNA template.
Termination
RNA polymerase reaches a termination signal (a specific DNA sequence), causing the enzyme to release the newly synthesized RNA and detach from the DNA.
What is translation?
the conversion of a sequence in mRNA (using ribosomes) into a sequence of amino acid
How many amino acids are there?
20
The mRNA sequence is made up of four types of _____ (A, U, G, C).
nucleotides
To make proteins, nucleotides are read in groups of three, known as ___.
codons
What is the universal start codon in RNA? in DNA?
What are the stop codons?
How many codons are there in total?
The remaining ___ codons code for the ___ different amino acids used to build protein
AUG (in RNA); ATG (in DNA) (1 codon)
UAG, UGA, UAA (3 codons; in RNA) TAG, TGA, TAA (in DNA)
64 codons in total
remaining 60 codons code for the 20 amino acids
What is a reading frame?
refers to the way nucleotides in mRNA are grouped into codons (triplets) during translation. The correct reading frame is necessary to produce the correct protein.
Correct Reading Frame:
AUG (Start codon) codes for Methionine (Met)
GCU codes for Alanine (Ala)
UGA (Stop codon) signals the end of translation
The protein sequence would be Met-Ala.
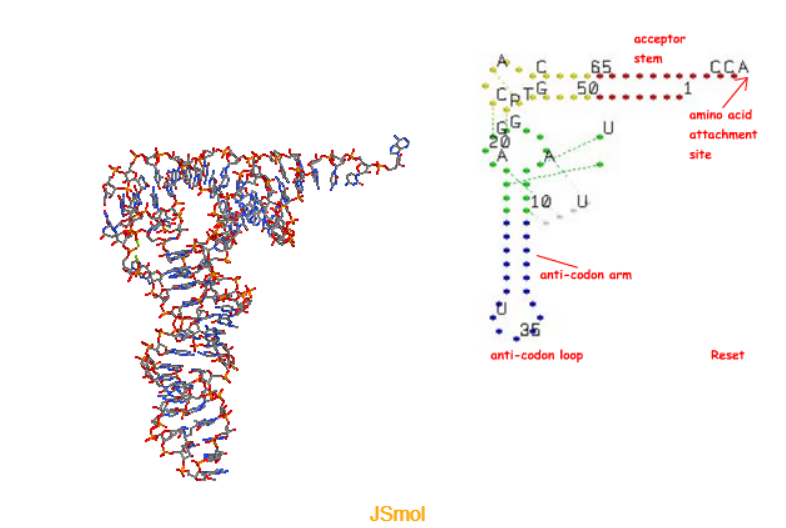
What are the structures of tRNA?
Acceptor stem: amino acids are attached to the 3’ terminus by the enzyme aminoacyl-tRNA Synthetases (aaRS). ~20-30 enzymes attach the correct amino acid to the correct RNA (aka charging the tRNA)
Anti-codon arm: The anticodon pairs with the codon through Watson-Crick base pairing, which involves hydrogen bonds between complementary bases.
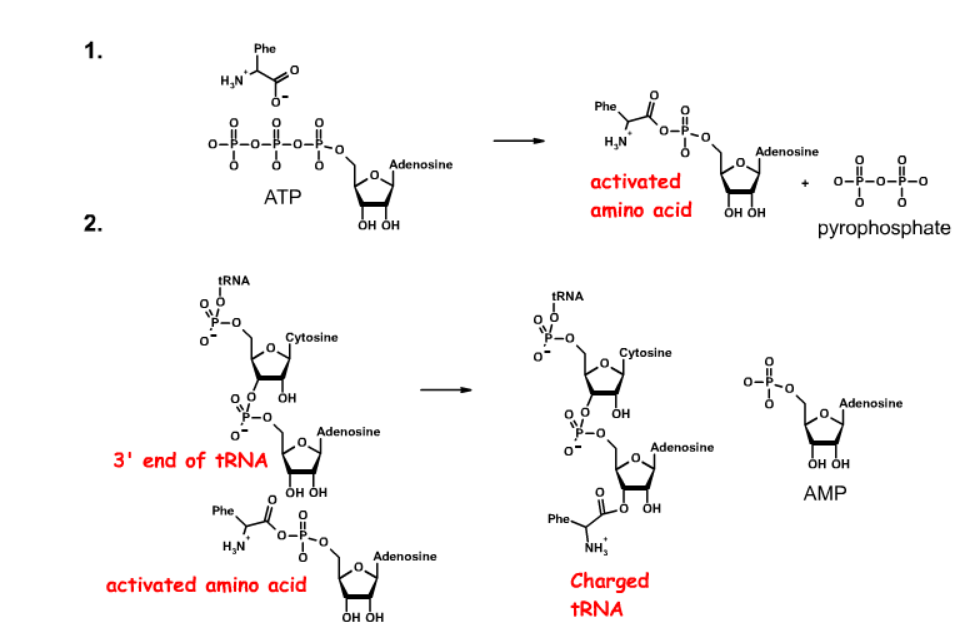
What are the steps of tRNA charging?
Charged by amino acyl tRNA synthetases
Step 1: The amino acid is activated by the attachment of an AMP to the carboxylate group. This reaction requires ATP and releases pyrophosphate
Step 2: he amino acid is transfered to the 3'-OH of the tRNA, releasing AMP
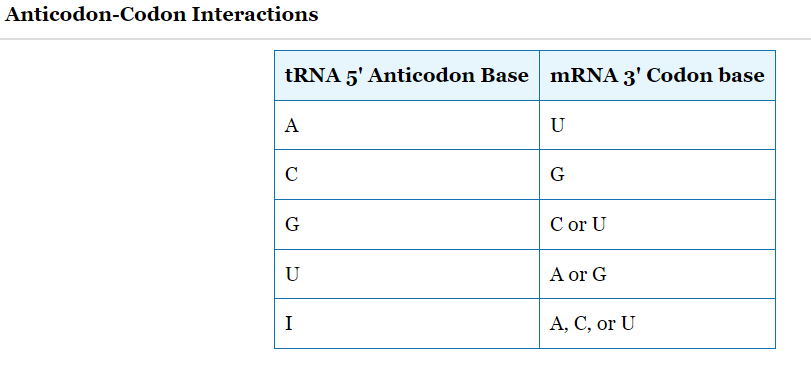
Codon-Anticodon Interactions
Charged tRNAs are selected by 1. _____ solely through codon-anticodon interactions.
Degeneracy at the 2. ____ position of codon-anticodon pairing allows multiple codons/tRNA.
The codon-anticodon pairing is 3. ______, as are most pairings of nucleic acid strands.
ribosomes
third
anti-parallel
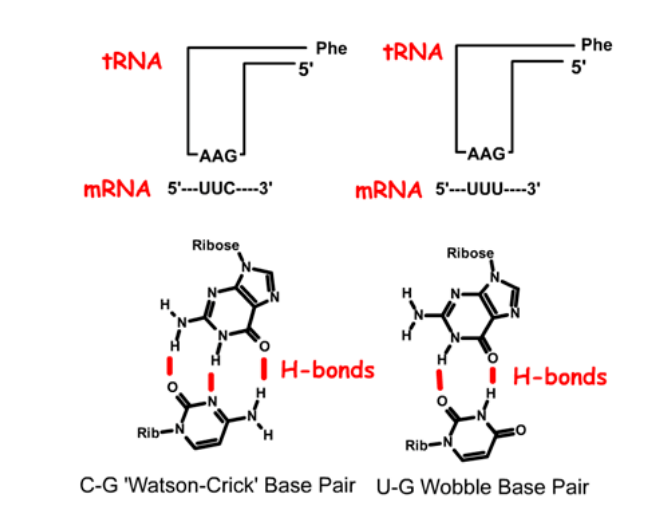
Pairing combinations for tRNAphe to the two Phe codons, UUC and UUU. In this tRNA, the anti-codon is 1. _____, or in reverse order: 2. ____'. Base pairing to the UUC codon uses standard Watson-Crick pairing. In the case of the UUU codon, the third base pair is a GU base pair, which is referred to as a "wobble" basepair.
5'-GAA-3'
3'-AAG-5'
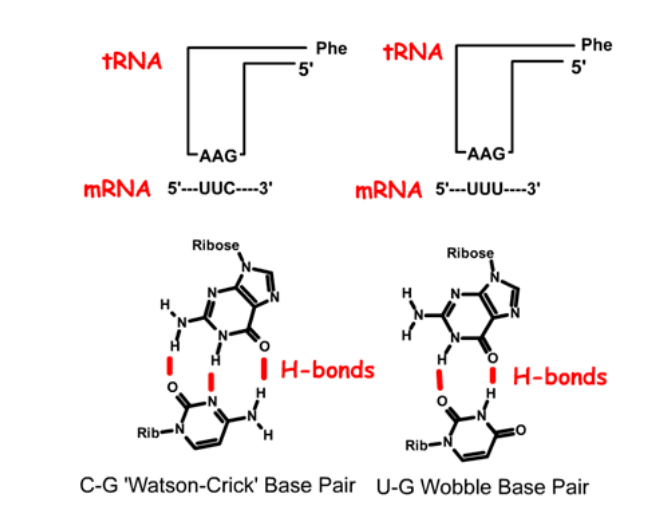
In anti-codon interactions the most versatile base is ____ which can pair with either A, C, or U in the third base of a codon.
It forms how many hydrogen bonds?
inosine
forms 2 hydrogen bonds
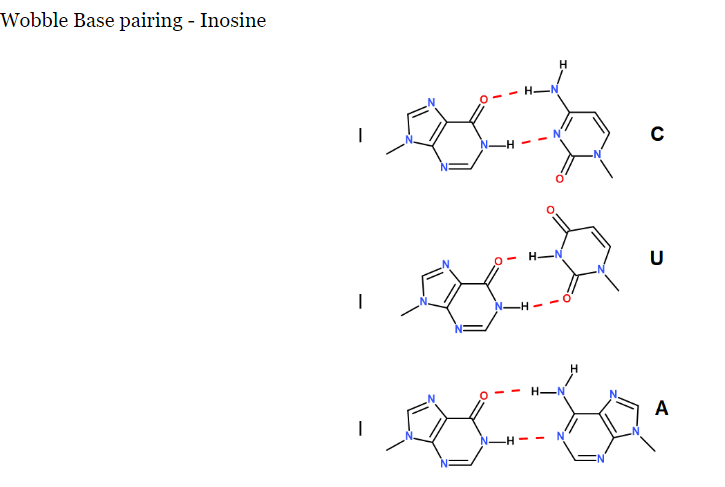

What are the pairings?
What is Sanger sequencing?
When you compare the mutant DNA to the normal (wild-type) DNA
It is used to identify sequence differences
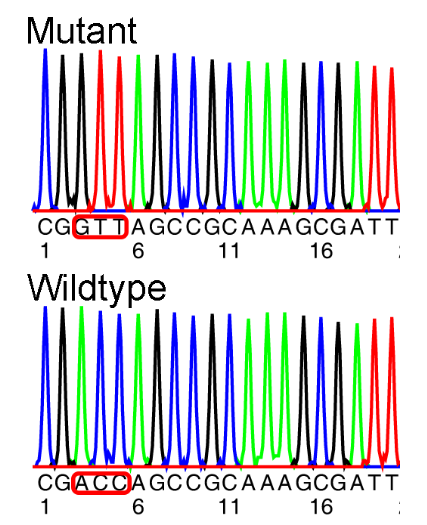
The changes in the DNA sequence can only be correctly translated if the ____ is known.
reading frame must be known
The correct reading frame starts with the third codon (ACC)

Which of the following conditions would cause the release of the Lac repressor protein from the lac operator site on DNA?
Presence of glucose in the growth media
Presence of lactose in the growth media
Presence of IPTG in the growth media
Presence of mannose in the growth media
Would NOT cause the release of the Lac repressor protein from the lac operator site on DNA
Would cause the release of the Lac repressor protein from the lac operator site on DNA
Would cause the release of the Lac repressor protein from the lac operator site on DNA
Would NOT cause the release of the Lac repressor protein from the lac operator site on DNA