Continuity & Change: D1.1 DNA Replication ONLY SL & D1.2 Protein Synthesis SL & HL
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
56 Terms
What is DNA Replication?
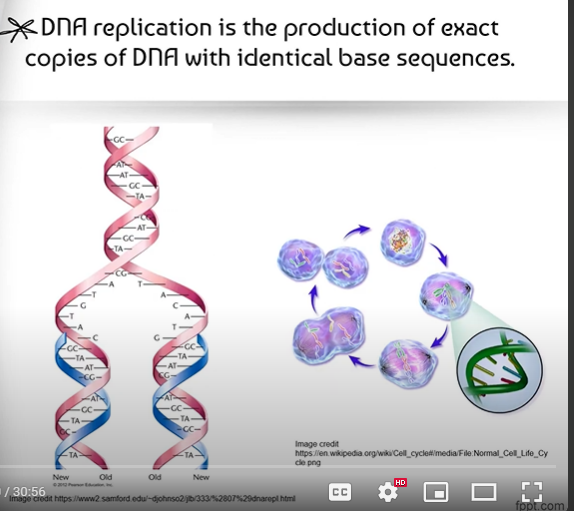
DNA replication is the process of creating two identical replicas of DNA with identical base sequences from one original DNA molecule.
What is the basic rundown of what happens in DNA Replication?
1. The original Strand of DNA is separated into two half strands.
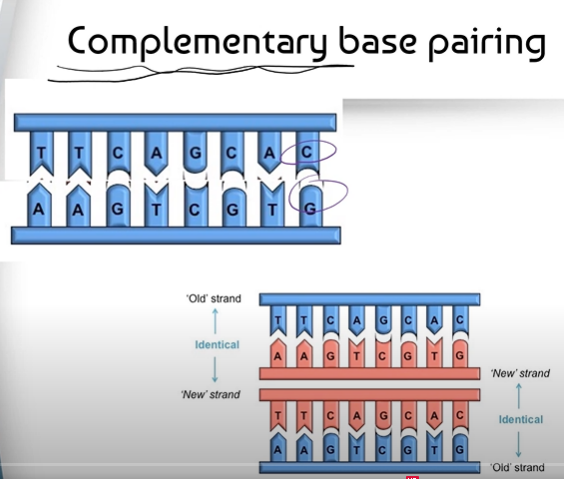
2. Complementary Base Pairing is used to add in new nucleotides to each of the two half strands of DNA. This creates 2 separate, identical strands.
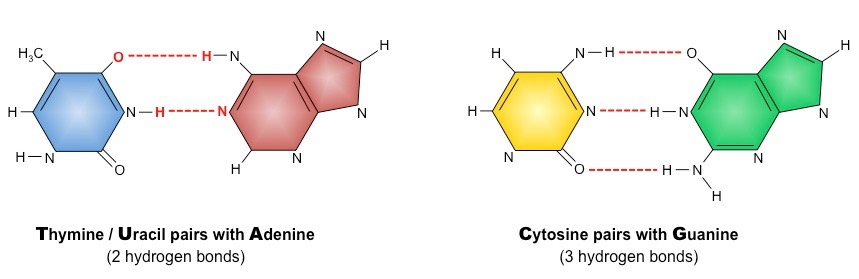
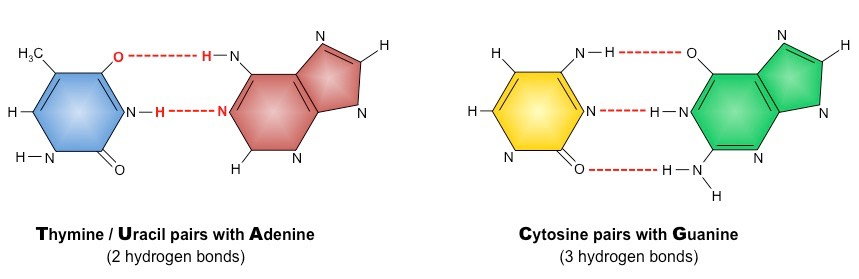
What is important about the structure of Base Pairs? How many hydrogen bonds hold together each set of complementary base pairs?
They have a structure that allows for the formation of hydrogen bonds. The base pairs Cytosine and Guanine are held together by 3 hydrogen bonds, while Adenine and Thymine is held together by 2 hydrogen bonds.
What does DNA Replication rely on?
DNA Replication relies on Complementary Base Pairing:
Cytosine - Guanine
Adenine - Thymine
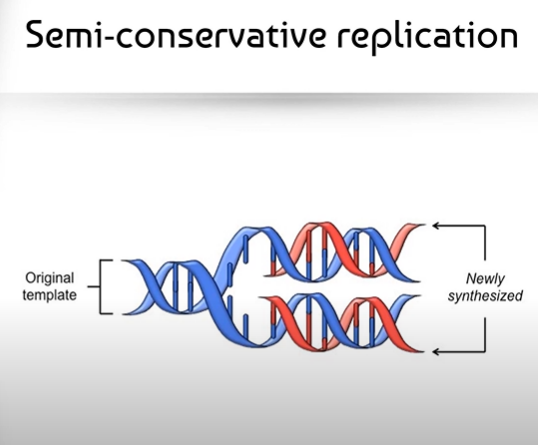
What is Semi-Conservative Replication?
In DNA Replication, the original strand is separated into two half strands, and each of these half strands form the template for new (free) nucleotides to attach, creating two identical double strands.
Each of these two identical strands contains half of the DNA from the original strand, and half from the newly formed strand; Therefore, we say it is semi-conservative, because we actually saved (conserved) half of the DNA in each new double strand.
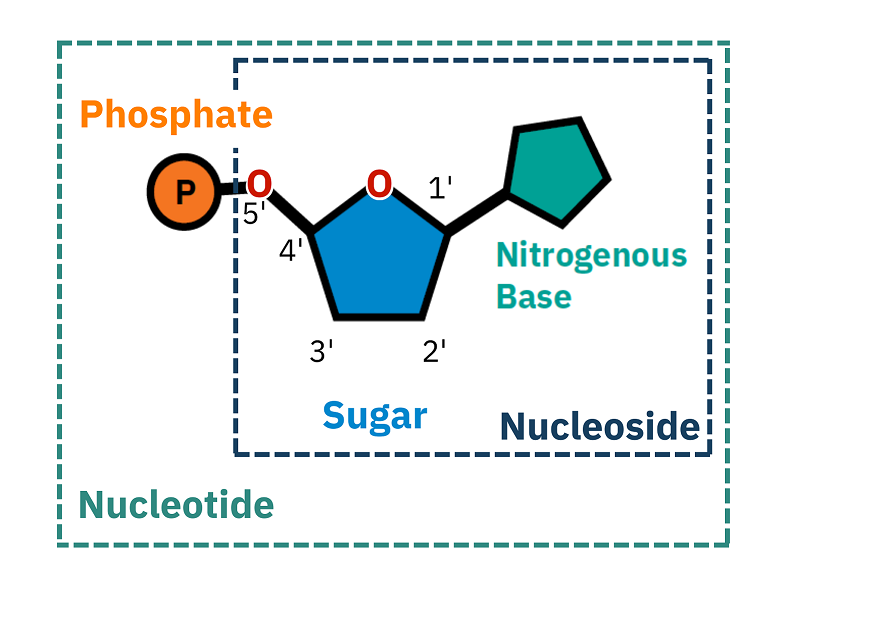
True or False: Are Bases and Nucleotides the Same thing? Distinguish between the two
No, nucleotides encompass bases; Nucleotides are the whole shabang; they include a phosphate group, a 5-carbon sugar (either deoxyribose or ribose), and a nitrogenous base.
"Bases" are just the nitrogenous bases in nucleotides, which are divided up into two types: Purines & Pyrimidines. Purines include Adenine and Guanine, and Pyrimidines include, Cytosine, Thymine, and Uracil.
What are polymers and monomers? What is the polymer & monomer in DNA Replication?
A polymer is a large molecule, and monomers are the small subunits that make up a polymer. In DNA Replication, the polymer is the DNA strand, and the monomers are the NUCLEOTIDES
What type of reaction takes place to form DNA strands in replication? What is used to catalyze these reactions? Where does DNA Replication happen?
Condensation reactions take place to form DNA strands, and enzymes are used to catalyze these reactions. DNA replication happens in the nucleus.
What is the liquid in the nucleus called? (where DNA replication happens) What is floating around in this liquid?
The liquid is called nucleoplasm. Free nucleotides not yet in DNA strands are floating around in this liquid.
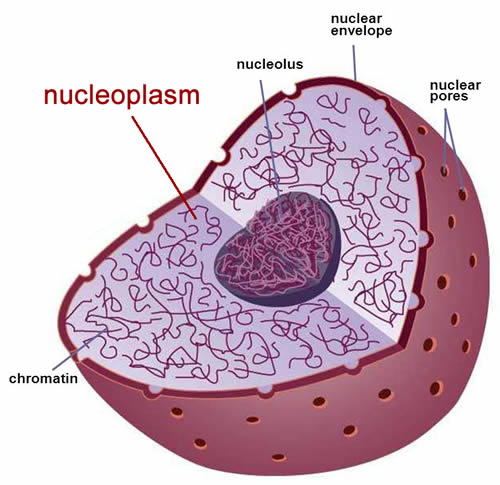
Why are the free nucleotides floating around in the nucleoplasm important for DNA replication?
The high concentration of free nucleotides allows complementary base pairing to occur naturally and spontaneously
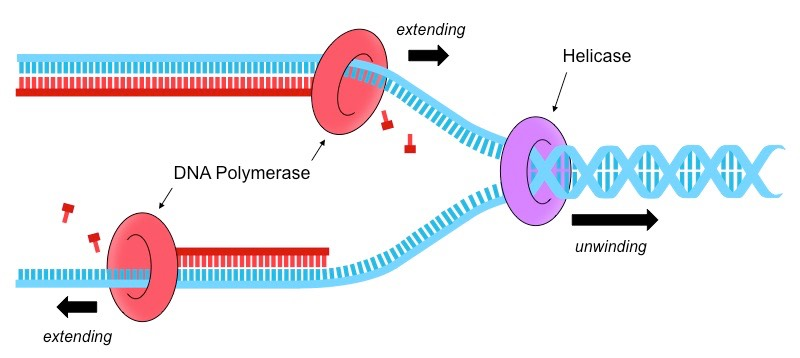
What are the names of the 2 most basic enzymes that help with the process of DNA Replication take place (SL only). Briefly explain the process of how they work together in DNA replication.
Helicase
DNA Polymerase III
1. Helicase opens up the double stranded piece of DNA, making it single stranded AKA Helicase unzips/unwinds the double helix of DNA, opening it up so that we have access to those nucleotides & so that complementary base pairing can occur
2. After helicase unzips & opens the double strand, complementary base pairs can slide into their new pairs and DNA Polymerase III will glue these new pieces of DNA using condensation reactions.
Is the process of DNA replication catabolic or anabolic?
Anabolic, because we are creating a big polymer of DNA out of small monomers known as nucleotides
What is the Polymerase Chain Reaction (PCR)? Don't explain the process, just what is it?
It is humans applying the process of DNA replication in Biotechnology.
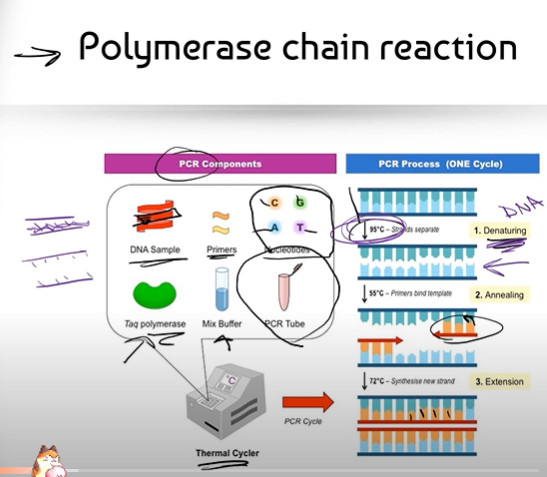
Can you break down the process of PCR (Polymerase Chain Reaction)?
(Part 1, before you explain the process of the Thermal Cycler)
1. We take a sample of DNA we want to copy (think of a small chunk of DNA that is double stranded)
2. We make some "primers" that are complementary to the beginning of the DNA sequence we want to copy
3. We also add in some free nucleotides that can complementary base pair to the DNA sample
4. We have the enzyme Taq Polymerase (that we stole from a bacteria called thermus aquaticus that lives in hot springs) that can glue those free nucleotides together, and a Mix Buffer, a solution that helps adjust the salinity and pH of the mixture so that the enzyme Polymerase can work at its best.
5. We put everything together into a small test tube called a PCR Tube
6. The PCR tube is then put into a machine called a Thermal Cycler. The Thermal Cycler's job is to change the temperature of the mixture in the PCR Tube.
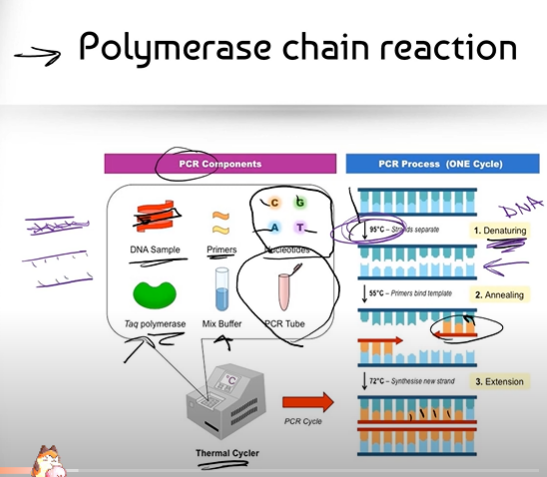
Can you break down the process of PCR (Polymerase Chain Reaction)?
(Part 2, explaining the process of the Thermal Cycler)
1. DENATURATION: The Thermal Cycler starts off by increasing the starting temperature of the PCR tube to about 95 degrees, almost boiling temperature. DNA is a double strand held together by hydrogen bonds in between its complementary bases; so when the temperature gets super hot, it denatures the DNA, causing it to wriggle/jiggle and become single-stranded
2. ANNEALING: Now that the strands are separated, there is room for the Primers to bind to the complementary bases on DNA. Annealing happens when the Thermal Cycler cools down to about 55 degrees, where the DNA strand is no longer moving/jiggling and the Primers can bind to the complementary bases.
3. EXTENSION: The Thermal Cycler heats the PCR Tube back up to 72 degrees, which is the optimal temperature for the (gluing enzyme) Taq Polymerase. Taq Polymerase can now glue together the nitrogenous bases: Adenines, Guanines, Cytosines and Thymines that complementarily bind to DNA.
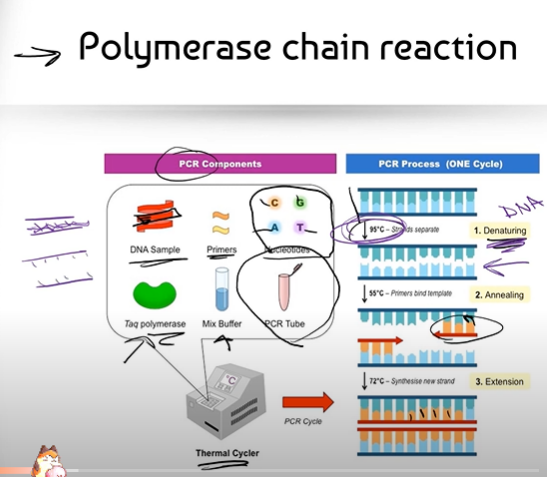
How long does the process of PCR take? Where does the name of the Polymerase enzyme used here come from?
The PCR process takes LESS THAN 2 MINUTES to complete; and it repeats to double the amount of DNA created.
The enzyme used is called Taq Polymerase because we stole it from a bacteria called Thermus Aquaticus that lives in hot water.
What is Gel Electrophoresis? Don't describe the steps, just say what it is
Gel electrophoresis is one of the ways we can analyze the DNA we replicated/copied in PCR.
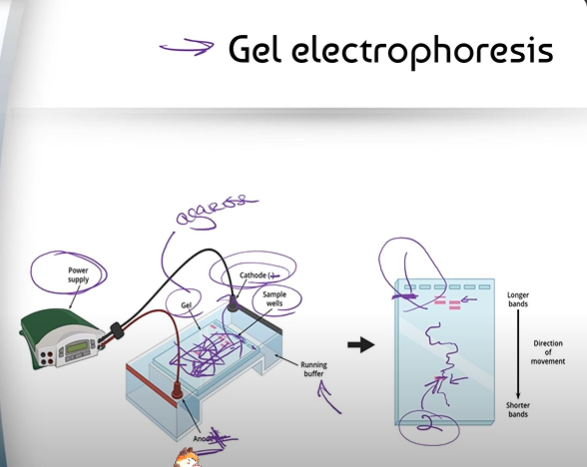
What are the different parts in gel electrophoresis?
In gel electrophoresis, there is a small block of gel, made of a substance called agarose
- some sample wells (holes) in the gel to insert DNA samples
- a running buffer (a lot of salt water solution to complete the circuit) underneath the small table of the device
- and a cathode + anode beside the gel, hooked up to a power supply
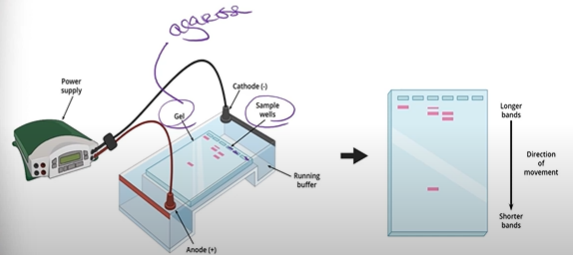
What is the process in gel electrophoresis?
DNA samples are inserted into the sample wells, and since DNA is negatively charged, it will be repelled by the cathode (-) and attracted to the positive side, the anode (+) on the other side of the gel. So DNA then migrates through the block of agarose gel towards the anode; but because the agarose gel is full of fibers, this makes it hard to move through, so as a result, little pieces of DNA move faster than big pieces. Note that this entire time, the anode and cathode are both hooked up to a power supply, which is helped by the running buffer solution to complete the circuit.
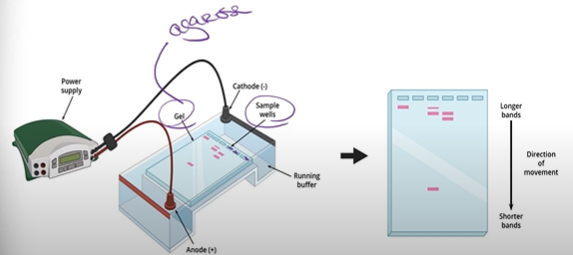
What is the result of gel electrophoresis?
you have different fragments of DNA suspended in the gel at different spots (because the smaller pieces moved faster than the big chunks of DNA), and now this pattern of DNA fragments can be analyzed and matched/compared with other fragments.
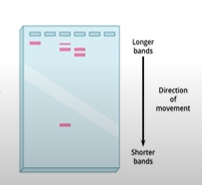
What is autoradiography (don't explain the process, just state what it is), and why is it called autoradiography?
it is taking pictures of the DNA fragment samples in our agarose gel. Autoradiography means self radioactive; the reason why will be explained in the next card
What is the process of autoradiography?
1. we make some of the DNA nucleotides in the agarose gel (from electrophoresis) radioactive so that they can emit their own radiation.
2. Now the DNA nucleotides themselves are radioactive, and can emit the radiation needed to take x ray film/pictures of them (without us shooting x rays at them).
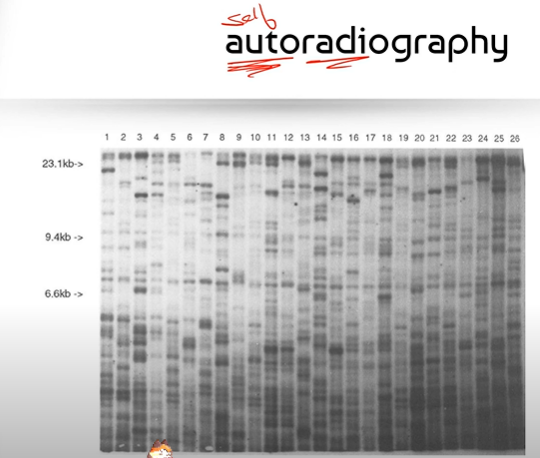
What do you generally see in an autoradiography picture?
you will see a bunch of little squares that represent a sample of DNA; the numbers at the very top indicate the # of samples of DNA, and measurements like 23.1 kB and 6.6 kB (measurements that decrease as you look further down the image) represent the length of each DNA fragment in kilobases, for ex. 23.1 kB is 23,000 bases long (kilo is 1000).
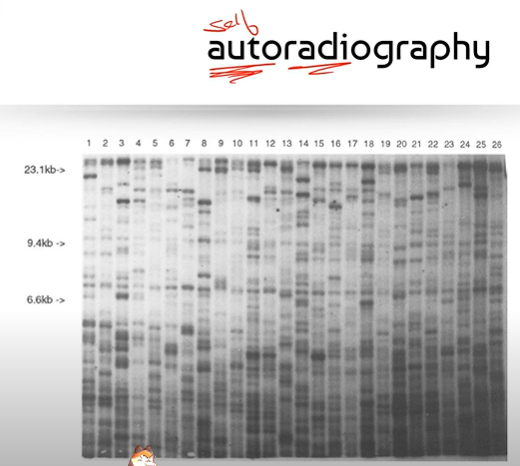
Why does the measurement in kB, of DNA fragments decrease as you look further down the image?
Because shorter DNA fragments move further in the agarose gel
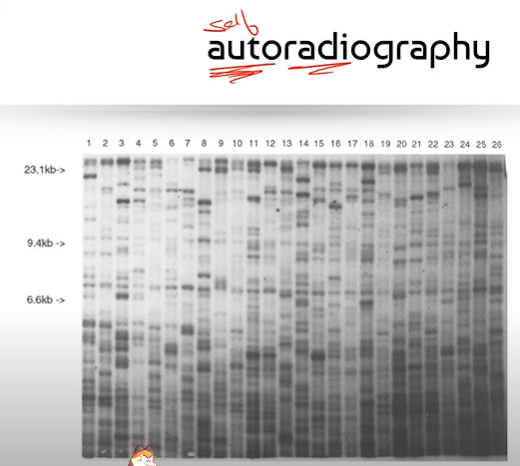
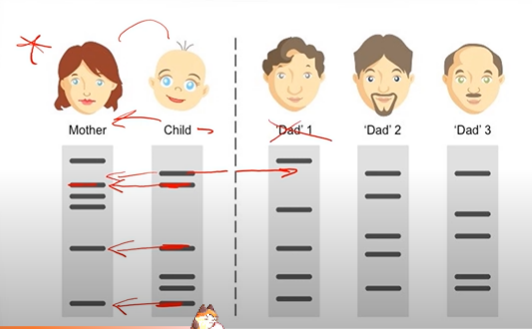
What is the point of using autoradiography? Explain one of these reasons.
To create images of fragment patterns that we can compare! This is especially useful for DNA profiling aka DNA fingerprinting in criminal cases or even paternity cases. In paternity cases, we take DNA samples from the mom, dads, and child, to determine which of the child's DNA bands were inherited from mom, and then determine which of the remaining bands not
inherited from mom correspond to bands that belong to one of the dads.
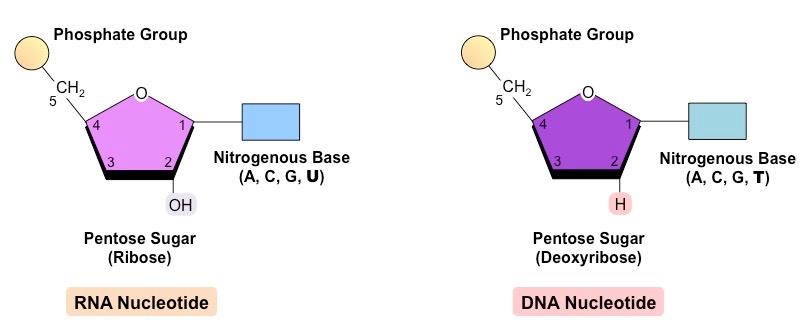
What does a nucleotide look like? In terms of its organic molecular structure when it is a part of the DNA polymer AND when it is just floating around?
Draw, label, and briefly explain why these two structures are different
When nucleotides are just freely floating around, they can have up to 3 phosphate groups attached. Why? Because there is a lot of chemical energy stored in these phosphate groups, and this allows the enzyme DNA Polymerase III to easily bind the nucleotides together during DNA replication.
Once nucleotides become a part of a DNA polymer, they only have one phosphate group.
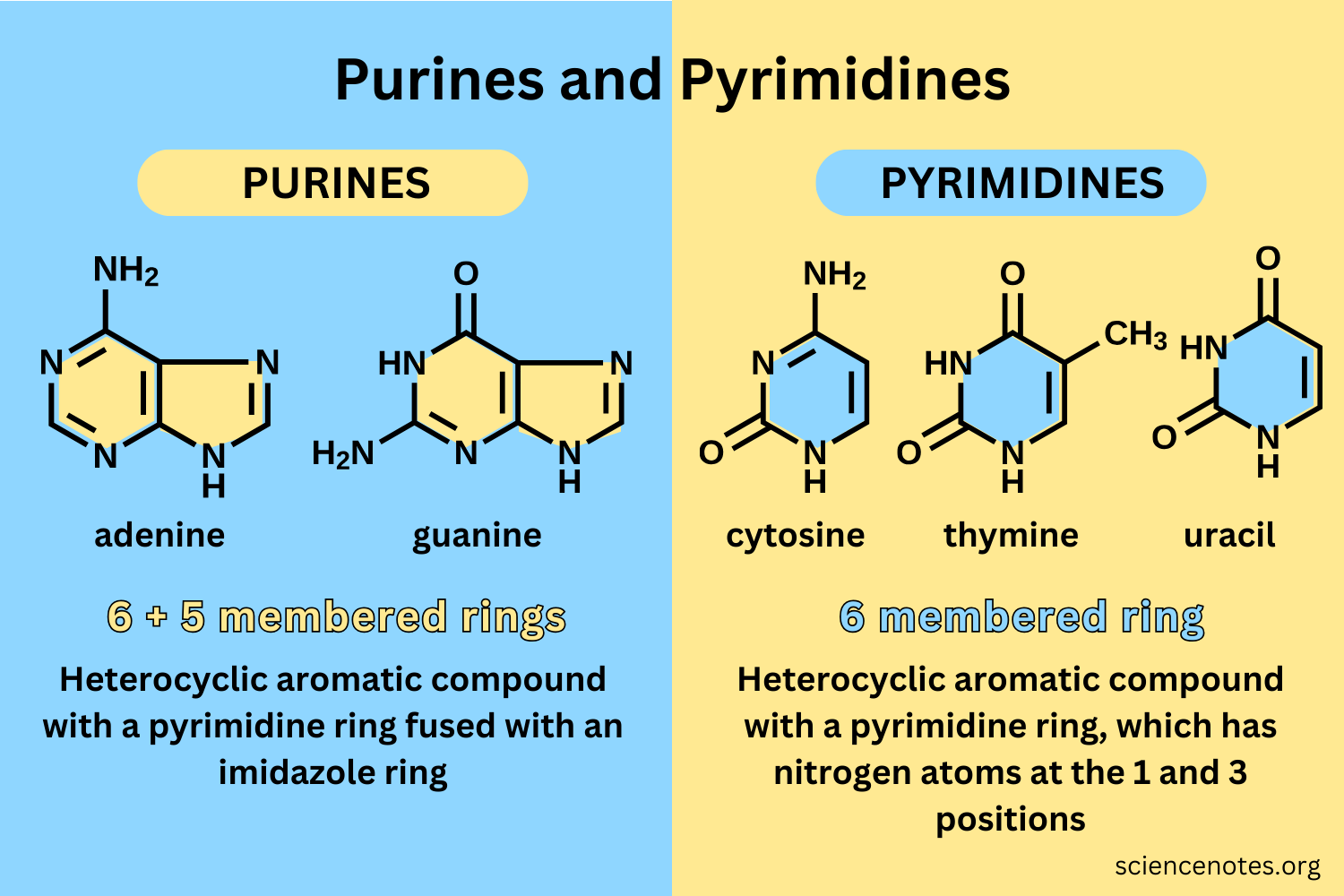
What is the acronym for remembering the classification of nucleotide bases (purines and pyrimidines)?
Purines (2 PUGA) = have two rings, only include guanine and adenine
Pyrimidines (Y CUT 1) = have 1 ring, include cytosine, uracil, and thymine
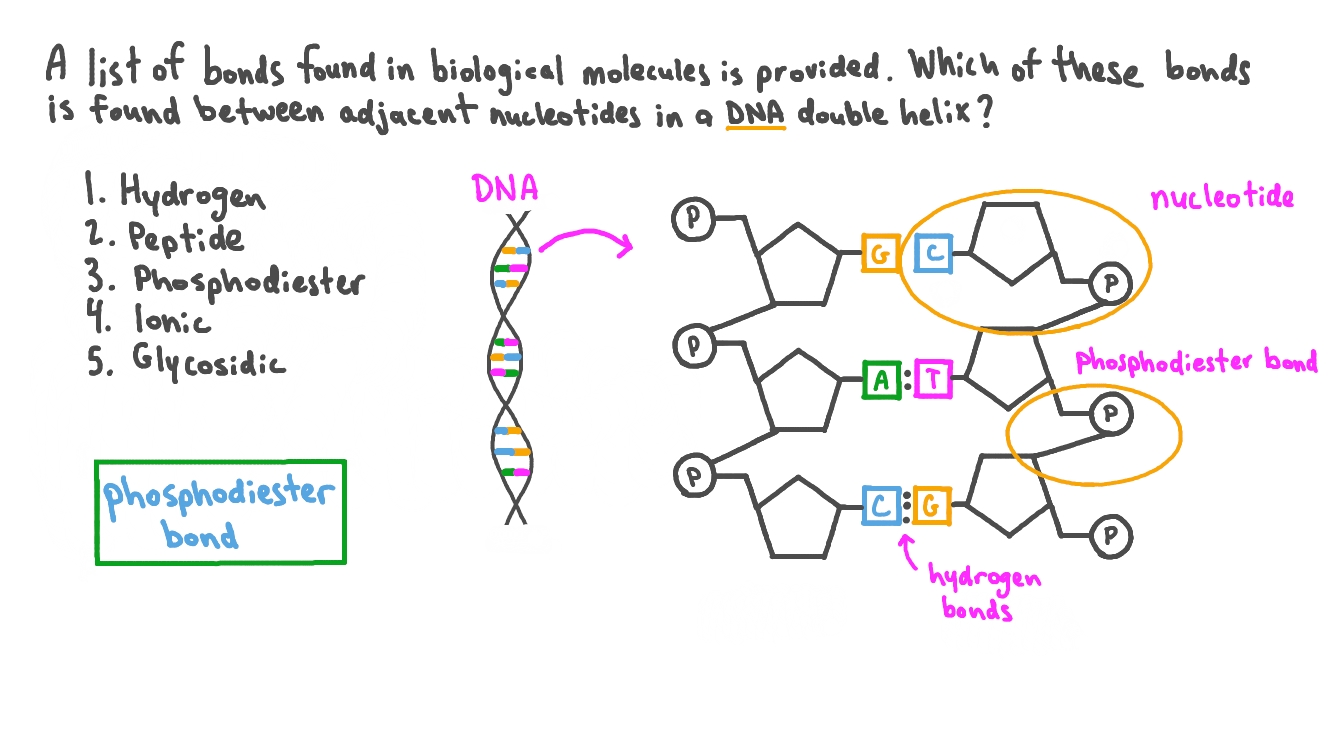
What kind of bond is formed between the 2 "glued" nucleotides?
A special kind of covalent bond known as a phosphodiester bond. The things being glued here are the phosphate group and the sugar.
What is "the Directionality of Replication" referring to?
It refers to the fact that DNA Polymerase III can only bind nucleotides together in a 5' to 3' direction (combined with the antiparallel structure of DNA strands)
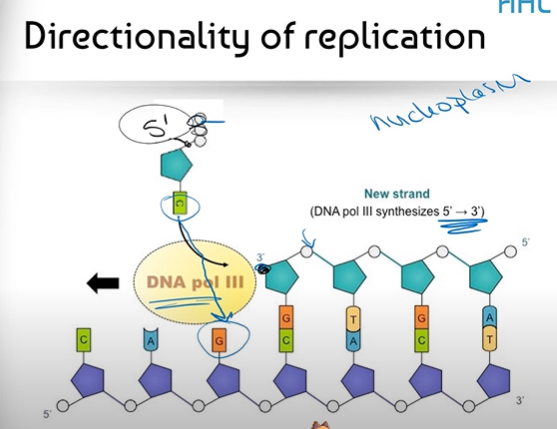
What does antiparallel mean?
It means that the structure of DNA runs in an opposite direction; so while one strand may have the 3' end on one side, the opposite end of that same strand will have a 5' end; and the strand opposite to it will run in the opposite direction
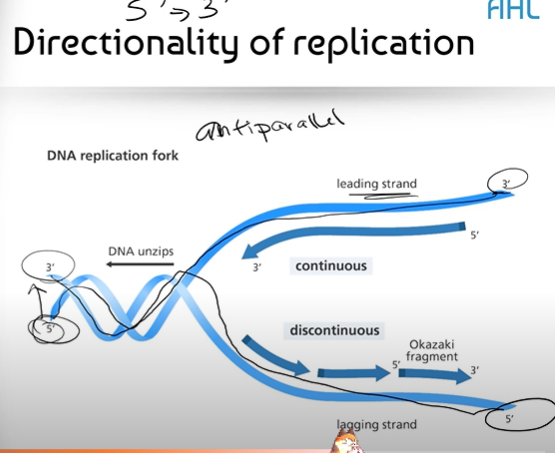
Can you try and draw the two strands of DNA in a double helix, labelling key parts involved in DNA replication?
In a DNA helix, you have the leading strand and lagging strand, the 5' and 3' end on either side, the replication fork, as well as a little arrow showing the direction that the DNA unzips in
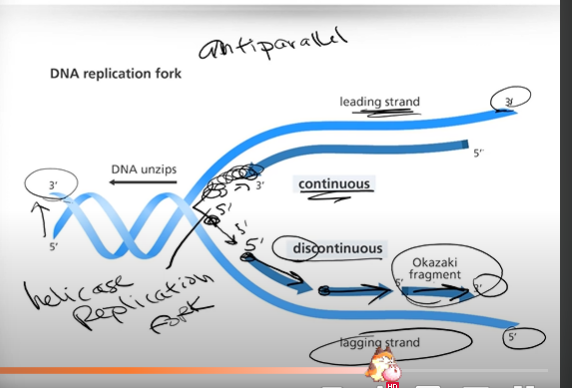
Can you explain what happens in the leading strand during DNA replication?
In the leading strand:
1. Helicase unzips/unwinds the DNA helix (in the direction of the replication fork)
2. This allows DNA Polymerase III to flow right behind the replication fork and continuously bind together new nucleotide bases (adenine, guanine, cytosine, thymine) along the DNA strand while Helicase continues opening up the DNA
*note!! this is all happening in a 5' to 3' direction, and the process of replication on the leading strand is called continuous replication*
Can you explain what happens in the lagging strand during DNA replication?
1. DNA polymerase III starts gluing nucleotide bases on the opposite side of the double helix (still in a 5' to 3' direction), and DNA is copied away from the replication fork.
2. Each of these little pieces are said to be copied in a discontinuous manner; And the fragments being copied in this discontinuous manner (on the lagging strand) are known as Okazaki fragments.
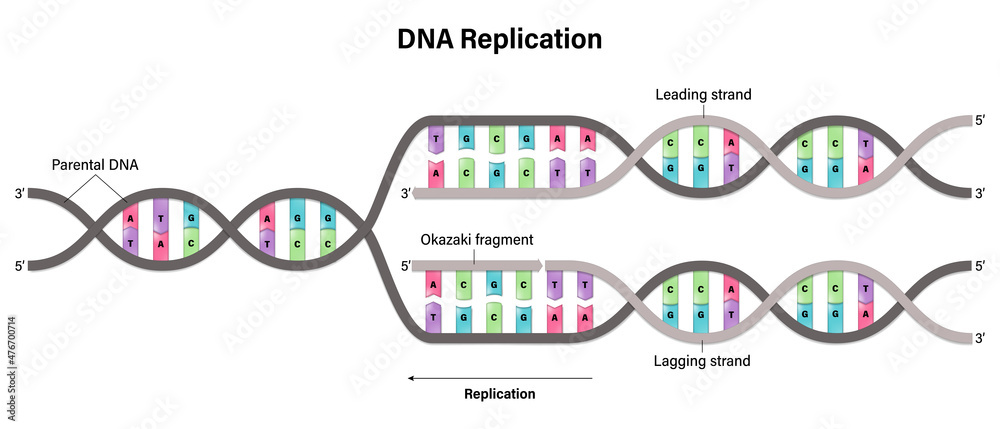
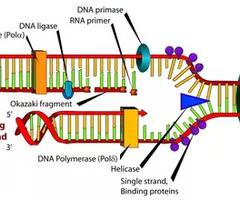
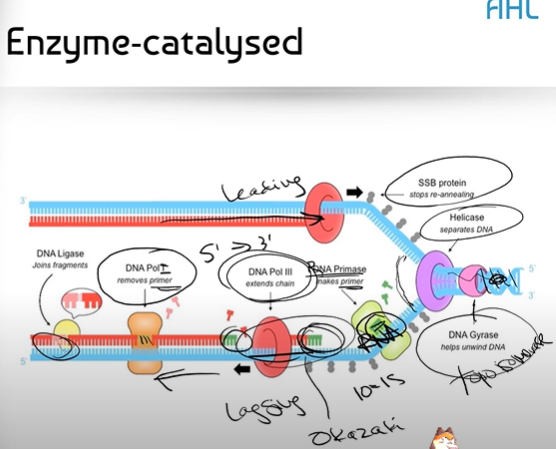
In detail, can you explain the function of each enzyme that helps catalyze the reactions on the leading and lagging strands?
1. DNA Gyrase helps relax the supercoiling of DNA, removing any twisty knots in the DNA so that Helicase can unzip it
2. Helicase separates DNA by unzipping/breaking the hydrogen bonds between complementary base pairs. As soon as helicase moved away, the complementary base pairs want to attract each other and zip back up again
3. SSB Proteins on the leading strand hold the DNA apart, preventing it from zipping back together (it stops re-annealing)
4. DNA Primase aka RNA Primase puts down a primer of about 20-15 nucleotides of RNA. We make a primer out of RNA because DNA Polymerase III needs a place to start. DNA Polymerase III cannot start if there isn't something already there.
5. DNA Polymerase III can now come in and bind together the nucleotides complementary to our base pairs in a 5' to 3' direction. (note: this process is discontinuous for the lagging strand, with Okazaki fragments, and continuous for the leading strand)
6. DNA Polymerase I removes the RNA Primer, filling in some of the gaps with DNA instead
7. DNA Ligase glues together these tiny gaps between the Okazaki fragments, left over from removing the RNA Primer
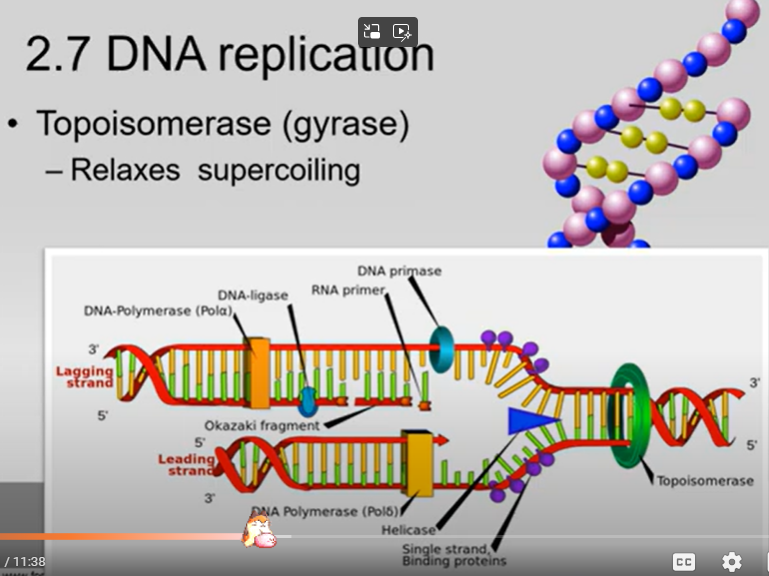
What is DNA Proofreading (just simply say what it is), and what enzyme is responsible for doing it in DNA replication:
DNA Proofreading is where DNA Polymerase III checks to see if any mismatches or mistakes were made between complementary base pairs.
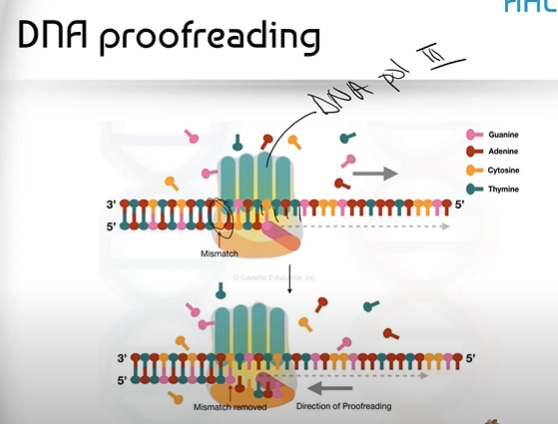
How does the process of DNA Proofreading actually work?
Before DNA polymerase III slides away from the DNA it just replicated, it squeezes the DNA to ensure that the width of it is just right. If it notices that something is off about the width, this indicates that the wrong nucleotide was put in, and so the very last chunk of DNA Polymerase III will go through a confirmational (shape) change to remove the wrong nucleotide and put in the correct one.
How often do mismatches happen in DNA replication?
Once every 10,000 bases, which is very frequent, considering that humans only have 3 trillion nucleotides)
What is the simple definition for gene expression, and what are the names of the two processes involved in gene expression?
Gene expression is the expression of specific genes; it is done through two processes: transcription and translation.
Define translation and transcription:
Transcription = using DNA code to produce a sequence of messenger RNA nucleotides (the messenger RNA (mRNA) transcript)
Translation = Taking the messenger RNA transcript and using it to put together a sequence of amino acids called a polypeptide chain. This polypeptide chain can then be folded into a functional protein.
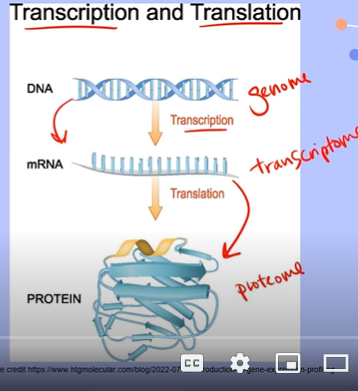
Draw 3 small pictures representing the general shape of the process of transcription, translation, and eventually the formation of the protein. Define the key terms involved in the processes.
Transcription: uses the Genome, which is the entirety of all genes
Translation: uses the Transcriptome, which is all of the messenger RNA created in Transcription
The formation of a Protein uses the Proteome, the collection of all proteins within a cell, tissue or organism
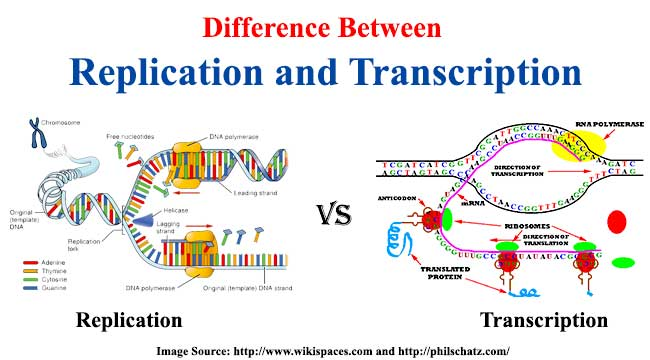
How is Transcription different from DNA Replication?
Transcription is the process of using a DNA template to create a messenger RNA transcript, while DNA replication is just the process of copying both strands of DNA to make more DNA. Also, DNA replication uses 7 different enzymes, while transcription only uses 1.
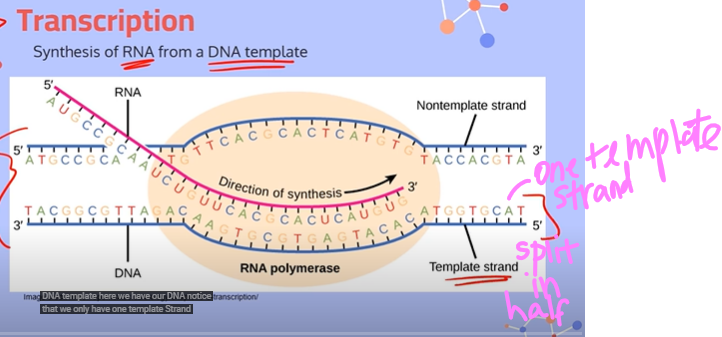
What are the strands used in transcription?
You take one complete DNA strand, break it apart in half, and then consider one half of the DNA strand to be the template strand, and the other half to be the non template strand. You also have a single stranded RNA that you use.
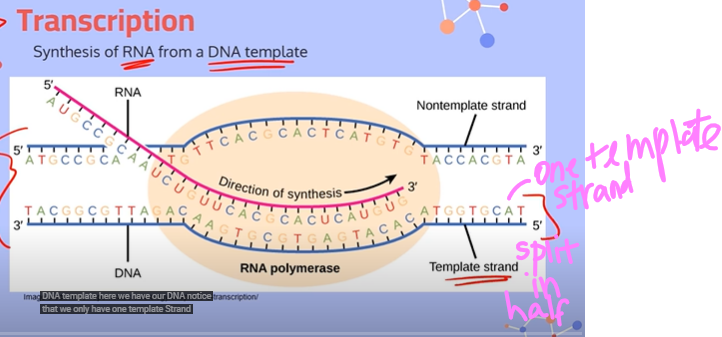
Explain the steps in transcription:
1. The enzyme RNA Polymerase will bind to a part of the DNA called the promoter region.
2. RNA Polymerase will then open up the DNA by breaking apart the hydrogen bonds between complementary base pairs. No helicase is required.
3. RNA Polymerase moves along the template strand, sliding into place the RNA bases complementary to the template DNA. It attaches these RNA nucleotides using phosphodiester bonds.
4. When RNA Polymerase reaches the terminator sequence, it will let go of the DNA strand to give us our piece of messenger RNA.
**note: everything goes from a 5' to 3' direction
How do nucleotide bases pair in transcription? What bonds hold these bases together?
One nucleotide base from the DNA and one nucleotide base from the RNA will be put together each time. Hydrogen bonds hold them together.
Guanine (of the DNA) + Cytosine (of the RNA)
OR
Cytosine (of the DNA) + Guanine (of the RNA) will still pair with each other
Thymine (of the DNA) will still go to Adenine (of the RNA), but not vise versa.
Adenine (of the DNA) cannot go to Thymine on RNA, because RNA doesn't contain thymine!!
**DNA only contains: Cytosine, Guanine, Adenine, and Thymine;
while RNA only consists of Cytosine, Guanine, and Uracil (but no Thymine)
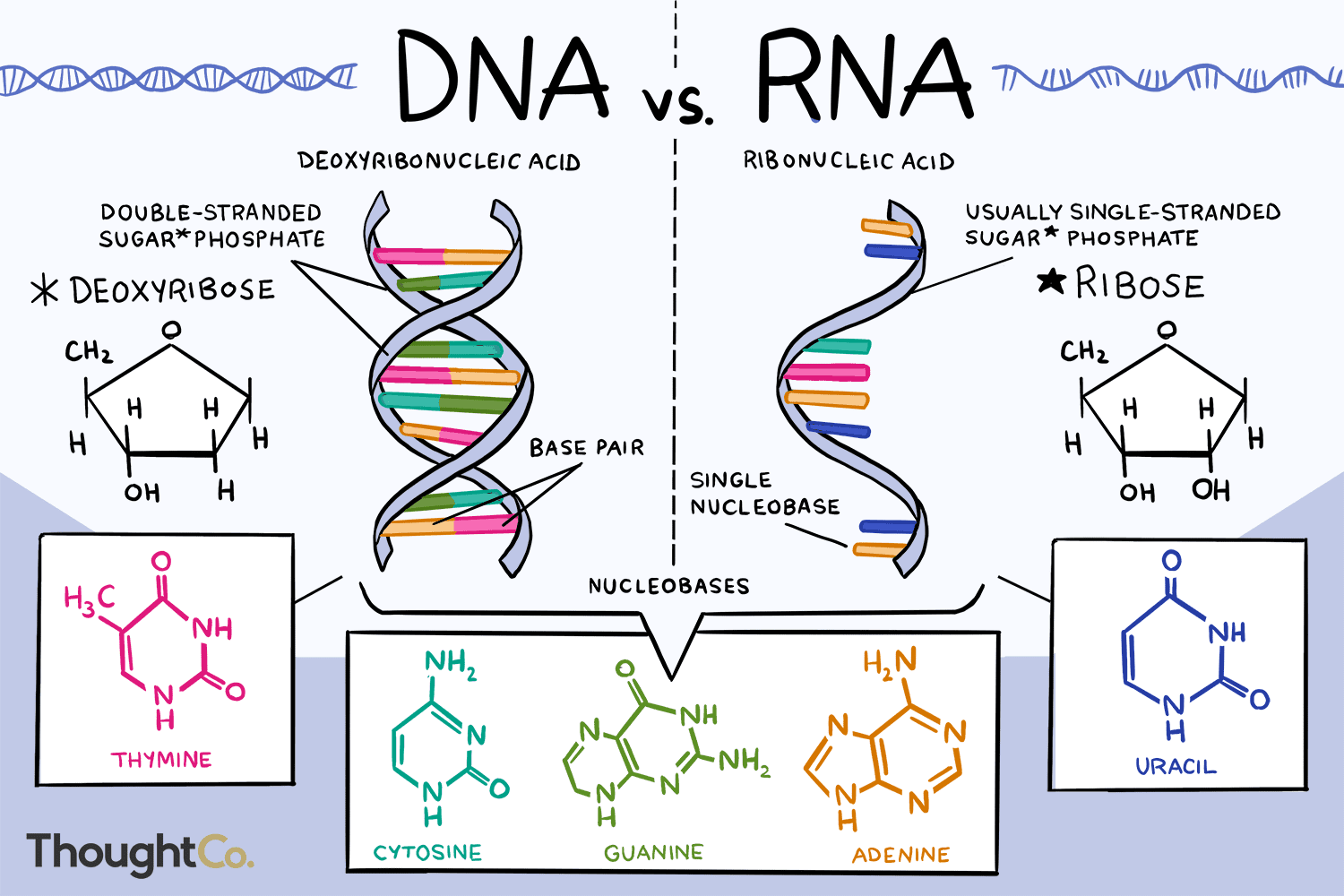
Contrast DNA Replication and Transcription:
DNA Replication:
- uses 7 different enzymes: Gyrase, Helicase, Primase, SSB Proteins, DNA Polymerase III, DNA Polymerase I, and Ligase
- the polymer created is DNA
- DNA is used to code for more DNA
- adenine + thymine, guanine + cytosine
- both strands of the DNA are template strands
- a primer is needed
- we copy the whole entire genome (so the entire genome is the template)
- DNA Polymerase III acts as its own proofreader
- There is no splicing in DNA replication
Transcription:
- only uses a single enzyme: RNA Polymerase
- the polymer created is messenger RNA
- DNA is used to code for mRNA (messenger RNA)
- base pairing is a bit different:
cytosine + guanine (interchangeable)
adenine (of DNA) + uracil (of RNA)
thymine (of DNA) + adenine (of RNA)
- only one template strand of DNA
- No primer is needed
- Only one gene is used to make messenger RNA (not the whole genome)
- There is no proofreading
- There is a process called splicing only in eukaryotes that allows it to modify pre-messenger RNA
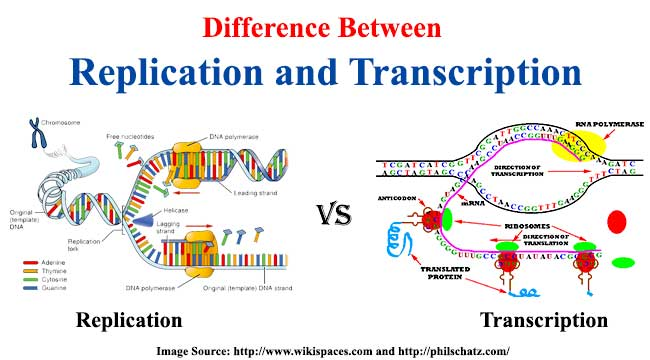
What are the similarities between Transcription and DNA Replication?
- both are controlled by enzymes, so both are metabolic in nature
- both are anabolic reactions (bc smaller monomers are being used to put together larger molecules)
- condensation reactions from phosphodiester bonds that hold the monomers of each polymer together
- both start with a DNA template
- both depend on hydrogen bonding between complementary base pairs
-both take place in the nucleus of a cell
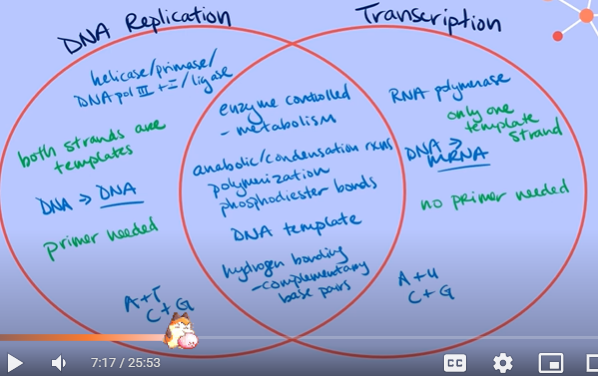
What is translation? (just briefly define it)
The process of using messenger RNA transcript to code for a sequence of amino acids that build a polypeptide chain.
These polypeptide chains can then get folded into a functional protein.
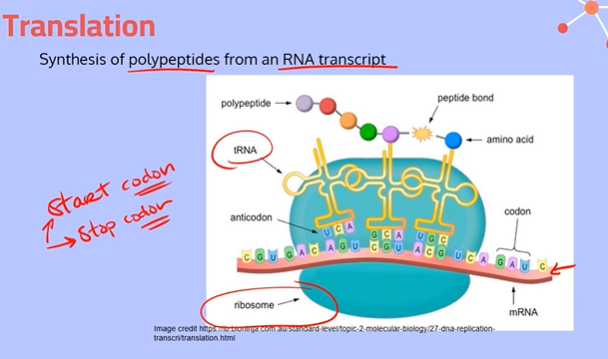
Where does the process of translation happen?
Translation takes places in the cytoplasm of the cell, specifically in the ribosome. (There are lots of ribosomes floating around in the cytoplasm)
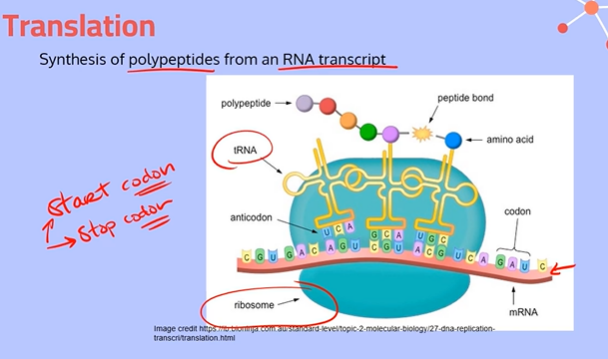
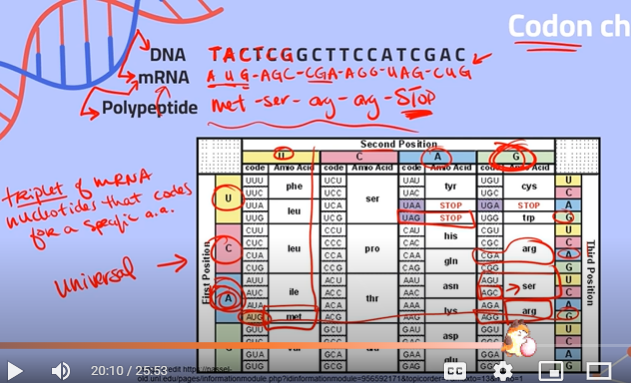
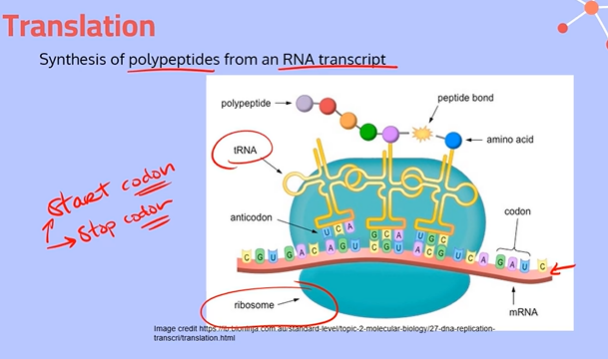
What are codons?
Triplets of mRNA nucleotides that code for a specific amino acid
What is the degeneracy of the codon chart referring to?
It means that multiple codons code for the same amino acid, because there are only 20 amino acids and about 64 different codons. This means you can have differences in DNA which won't impact the protein coded at the end of translation.
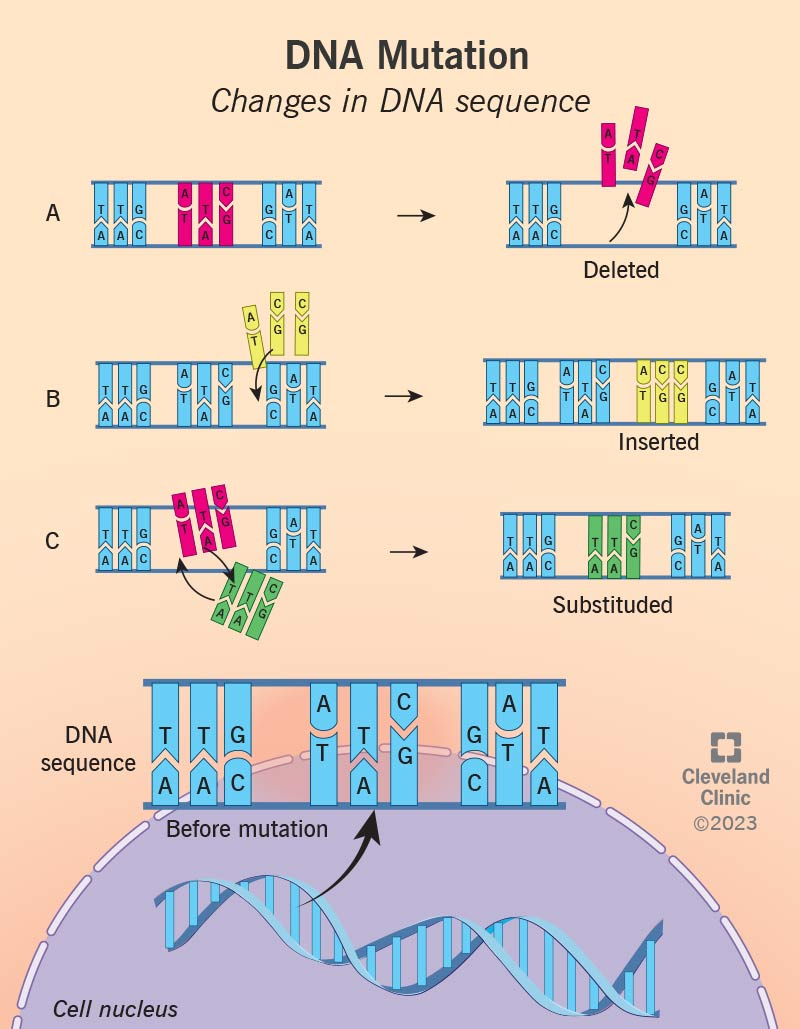
What are mutations? What are gene mutations evidence of or what are they a result of?
Changes in the DNA sequence of a gene.
It is a result of the degeneracy of the codon chart. Even though we said that codon degeneracy allows us to have changes in DNA sequencing without impacting protein structure, this isn't always true; since some changes in DNA sequences can lead to significant changes in protein shape, and as a result, protein structure. This is what we call mutations.
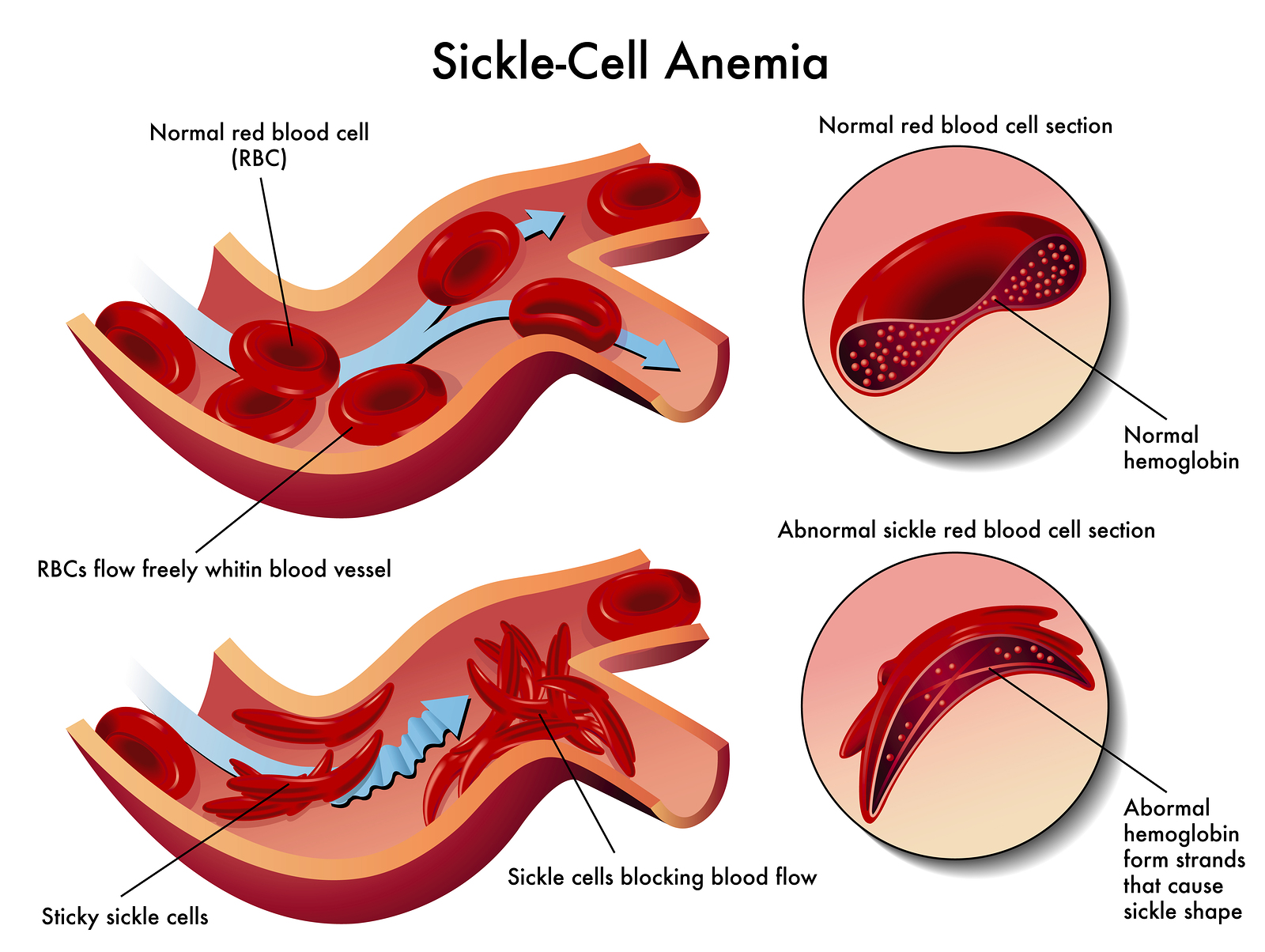
What is one kind of mutation?
Sickle cell anemia, a base sequence mutation that causes the shape of hemoglobin (especially in environments with low oxygen) to morph from concave red blood cell shapes to sickle shapes that will clot in very small capillaries, causing tissue damage and incredible pain.
What are the names of the 7 different enzymes that help catalyze DNA replication? Draw a diagram to show them.
1. DNA Gyrase
2. Helicase
3. SSB Proteins
4. DNA Primase
5. DNA Polymerase III
6. DNA Polymerase I
7. DNA Ligase
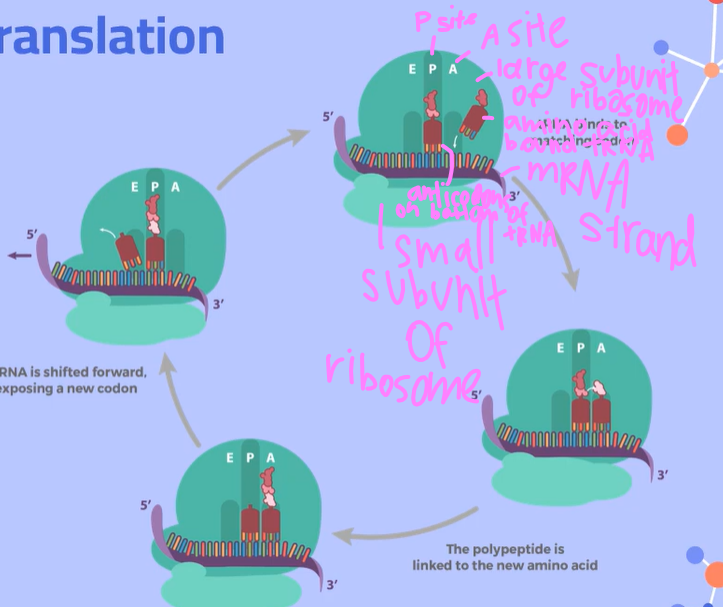
Explain the process of translation, going into depth about the different Sites
Initiation:
A ribosome and initiator tRNA attach to the start codon of an mRNA molecule (messenger RNA molecule). The small and large subunits of the ribosome also bind to the mRNA.
2. Elongation:
At the end of each tRNA molecule (transfer RNA molecule), there is an anticodon, which is complementary to the codons on the mRNA. These codon and anticodons pair together using hydrogen bonds between complementary bases.
The tRNA molecule brings in an amino acid from the cytoplasm. When it reaches the ribosome, the tRNA starts at the P Site with its amino acid.
Then a second tRNA carrying another amino acid will slide into the A Site. The large subunit of the ribosome catalyzes a reaction, forming a peptide bond between the newly arrived amino acid and the amino acid held by the tRNA in the P site. The ribosome transfers the entire growing polypeptide chain from the tRNA at the P site to the amino acid on the tRNA at the A site.
Now the tRNA in the P site is empty and uncharged, so it slides over to the space in the E site and exits the ribosome to pick up another amino acid. The tRNA in the A site, now with the growing polypeptide chain, is going to slide over to the P site; This process repeats until the ribosome gets to a stop codon.
Compare and contrast transcription and translation:
TRANSCRIPTION:
carried out by the enzyme RNA Polymerase
we form:
an mRNA transcript (which is our polymer)
nucleotides make up our mRNA transcript (monomers)
phosphodiester bonds are formed between these monomers
DNA is converted into mRNA
does not require tRNA
complementary bases are 1 DNA to 1 RNA base
TRANSLATION:
ribosomes catalyze the formation of chemical bonds
we form:
-proteins (polymers)
-amino acids (are the monomers that make up the polypeptide chains in the protein)
mRNA is converted into polypeptide chains
mRNA is converted into polypeptide chains
mRNA bases are complementary to tRNA bases
triplets of mRNA bases are called codons
complementary to these mRNA bases are the triplets of tRNA bases called anticodons
BOTH:
both are metabolic reactions
both are anabolic
both use condensation reactions
both involve mRNA
both involve hydrogen bonding between complementary base pairs
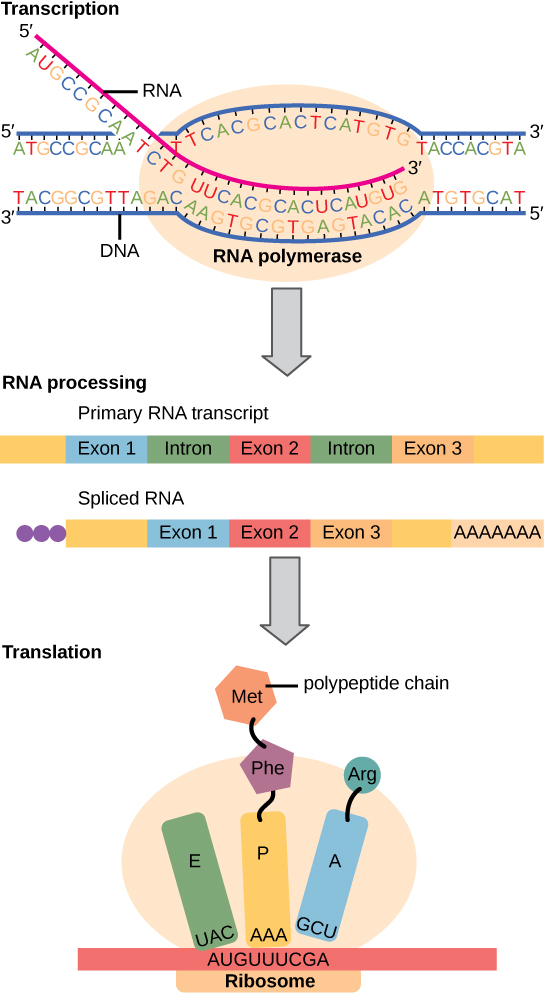
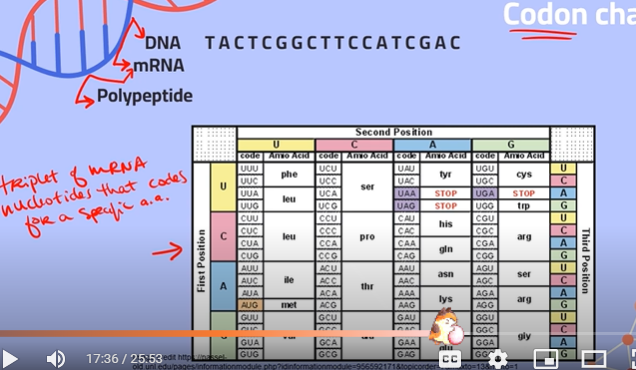
Find the corresponding mRNA sequence and the amino acid produced:
**recall:
thymine (of DNA) —- adenine of (RNA)
adenine (of DNA) —- uracil (of RNA)
cytosine (of DNA) —- guanine (of RNA)