GENET 390: Topic 2.3 - PCR
1/108
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
109 Terms
What is the objective of PCR?
Amplify DNA sequence that is between + including 2 Primers using ANY template DNA
What are the basic overarching steps to PCR
3 Step Cycle
Denaturation
Annealing (primers)
Elongation
****What are the Temperatures associated with each Step of the cycle?
Denaturation = 94
Annealing = 50-60
Elongation = 72
What is a HOT START?
TOTAL Denaturation of template DNA
Denaturation cycles of 94 for up to 5 minutes
****Why is a Hot start CRUCIAL for successful amplification?
Avoids MISPRIMING
ensures the DNA synth only occurs when temps are close to Tm to ensure high STRINGENCY/Specificity
***Define MISPRIMING
Annealing of primers to Incorrect locations during the ramp up of temp from Room temp to Denaturation temp
b/c The lower temperatures allow for mismatch (low temp = low stringency)
****Why is MISPRIMING an issue?
Polymerization can occur at any time
if the wrong sequence starts polymerizing all future cycles will contain the misprimed sequence
Amplification of UNDESIRED products
***What are the 4 possible methods for the Hot Start to ensure Mispriming does not occur. Indicate which one(s) are always used.
Everyone at least uses this one
Make entire PCR Master mix on ICE + transfer to Thermocycler already at 94
Additional Quality control if the first one doesn’t work
Don’t add Taq until after hot start
Separate Taq from rxn mixture using a thin wax layer
Try Diff primer
***What is a PCR Mastermix
Everything needed EXCEPT Taq
****What is Taq?
Taq = the thermostable Polymerase that will elongate the DNA during PCR
What is the Thermocycler?
The PCR machine that does all the temp stuff
*****What are the steps of a PCR cycle again? What are the approx. lengths of time each step takes + what factors does the exact time depend on?
Cycle = 95/94 —> 55 —>72
Denaturation (94) = ~ 1-2 minutes
Time depends on a couple variables
Annealing (50-60) = ~ 1-2 minutes
Depends on PRIMER
Elongation (72) = ~1-5 minutes
Time reflects LENGTH OF DESIRE PRODUCT
****Why should Elongation time Reflect the length of the Desire Product?
The polymerase (Taq) replicates at a known rate
about 1kb/minute
amount of time given will dictate length of product made
****Why is Elongation Temp 72?
Taq works best at 72
***Which cycle is the most important cycle?WHY?
FIRST
All future cycles = based off of products of the first
****Is the desired product made after the First cycle?
No
***What is the product of the First cycle? WHY?
An extended product
Taq extends off primers until it FALLS OFF = more than needed (~6-7 kb)
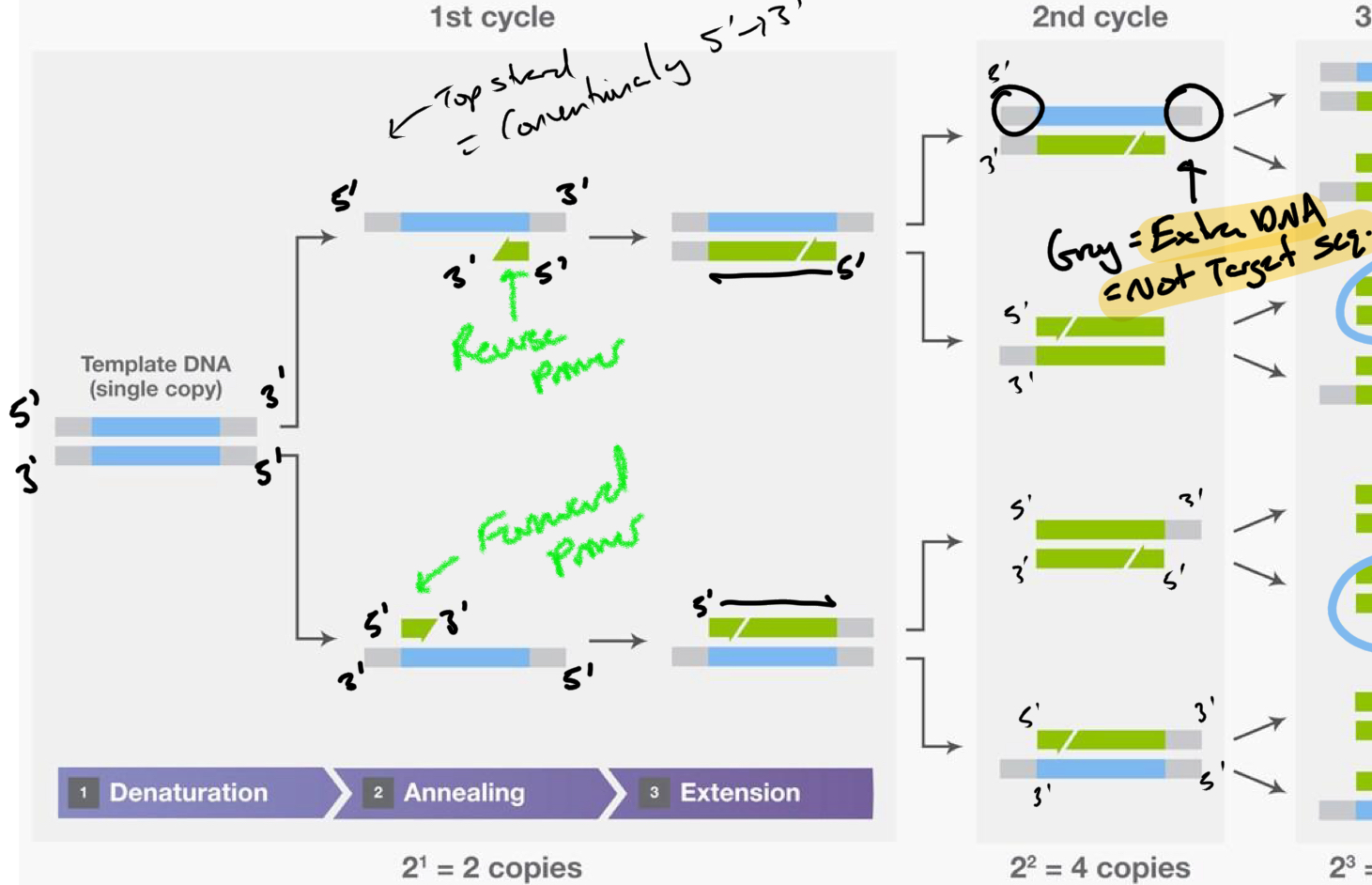
***In which cycle is the Desired product first made?
Cycle 3
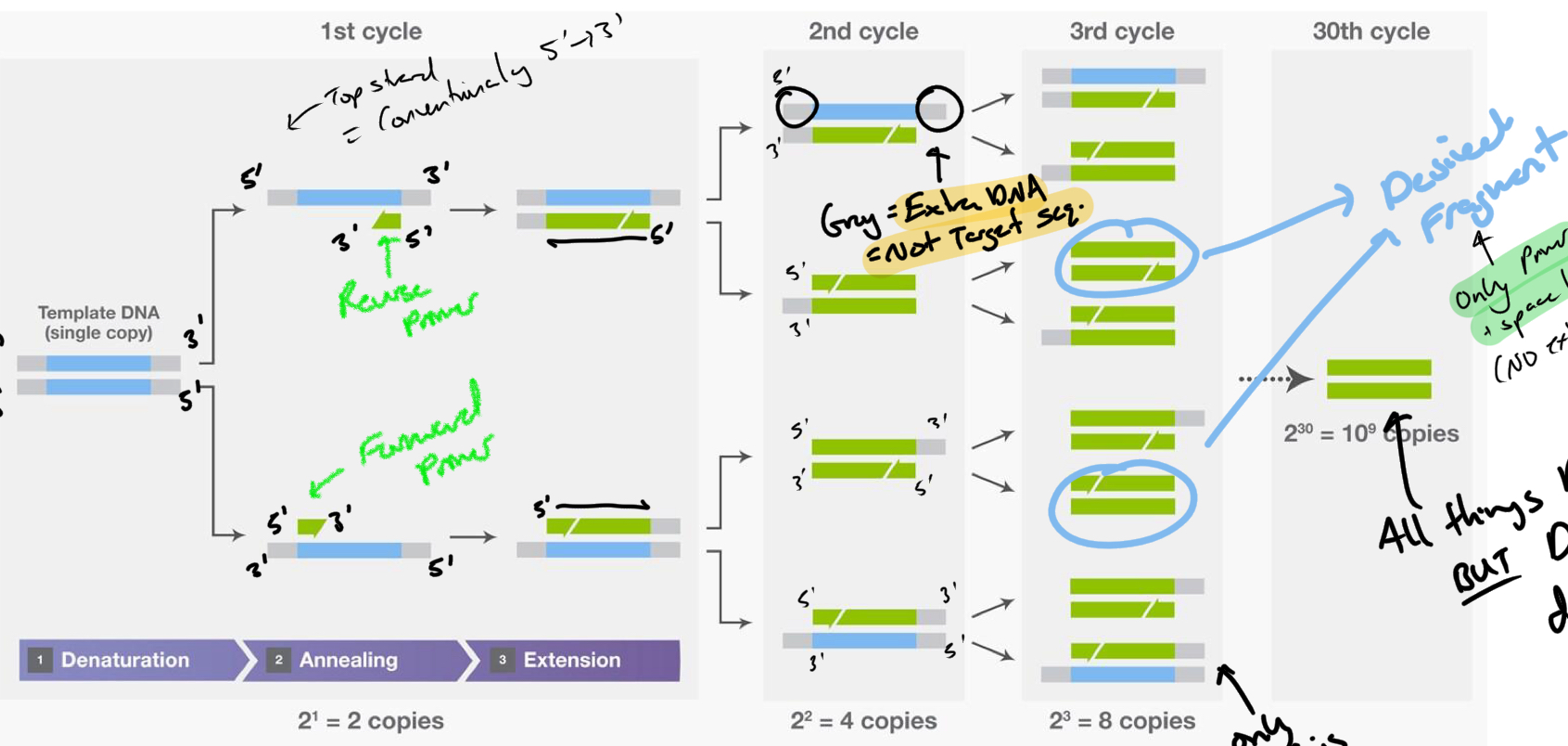
****What is the Product(s) of cycle 2?
DNA made from Extended strand from product 1
There is still extra unneeded DNA from the template(product 1)
+ DNA same as product 1
****Other than the Desired Product, What other products are present in CYCLE 3?
Products of Cycle 1 + 2
****If all things remain in the tube between each cycle, WHY does the DESIRED PRODUCT DOMINATE after the remaining cycles finish?
EXPONENTIAL GROWTH of desired/full length product
Linear amplification (arithmetic) of Extended products (cycle 1 + 2 products)
What is the desired product composed of
Primers + Space between the Primers
NO extra DNA
Finish the sentence: Primers allow for _______ in what gets amplified
Specificity
How many times do we repeat the cycles? Why?
~ 30 times
After 30th cycle AMPLIFICATION WILL PLATEAU = useless
****What are the 5 REASONS for why amplification will Plateau?
Substrate limitations
Primer-dimer formation + accumulation
dNTPs degrade
Taq progressively degrades
Accumulation of Products
***Explain reason 1. Substrate Limitations. Why does it occur + cause plateau?
Only so much of each starting ingredients can be added + eventually run out
***Define primer dimers
base paring between different primers
***Explain reason 2. Primer-Dimers. Why does it occur + cause plateau?
Once all the ingredients are almost used the excess PCR cycles promote self and cross-annealing between primers and produce dimers.
***Explain reason 3 + 4 dNTP + Taq Degradation. Why does it occur + cause plateau?
Heat denatures everything. Both can only withstand ramp up to 94 so many times.
If the building blocks and machinery are broken product can no longer form
***Explain reason 5. Accumulation of product. WHY does it cause plateau?
High DNA concentrations causes faster reannealing because reannealing is concentration dependent
higher concentration = more likely complementary strands will bump into each other = denaturing harder and renaturing easier
Finish the sentence: Primers represent the ____ point and _____ point of a PCR reaction
Start point and end point
*****At least HOW LONG should primers be? What is the MAX LENGTH? WHY?
20 ntd for specificity MAX = 30 ntd to preserve specificity
Too short = too easy to find match that might not be right.
Too long = hard to find a perfect match
Are primers dsDNA or ssDNA?
ssDNA
*****What are the 2 PRIMERS where do they anneal and how are they each related to the Coding strand?
the FORWARD + REVERSE primers flank sequence of interest
Forward Anneals on the Left of the NON-CODING STRAND (seq = same as coding strand)
Reverse Anneals on the Right of the CODING STRAND (seq = complementary to coding)
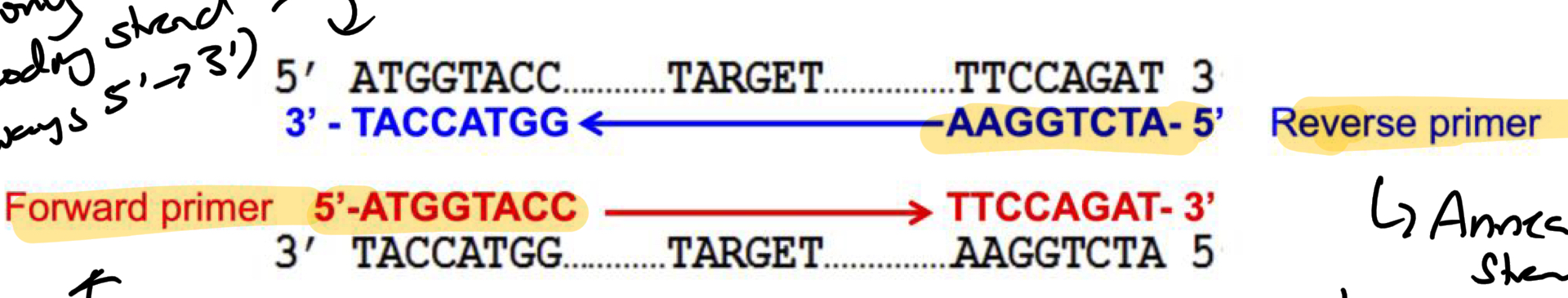
How is the Coding strand always shown? 5’ —> 3’ or 3’ —> 5’?
5’ —> 3’
*****Where is the 3’ end of the Primer always pointed? Why?
IN
Polymerase needs a 3’ end for polymerization
****True or false: Primers are not included in the final product?
FALSE
*****Other than length what are 5 OTHER factors you need to consider when designing primers?
Tm of the primers
G/C content
Homopolymer runs
GC rich 3’ end
Complementary sequences
What are the Tm requirements for a primer? (3 aspects) Why?
Tm’s of the 2 primers should match
anneal around the same temp (5 degrees max difference)
Tm’s should NOT be higher than 50-60 degrees
Too close to 72 degrees = would not anneal enough to have proper extension
Need to anneal to target before template reanneals
****What should the GC content be between? What should it always be less than and why?
GC between 40 - 60%
strengthen base pairing between primer + template
Keep Tm at a temp above risk for FALSE PRIMING
Always less than 70%
To prevent too high of an annealing temp close to 72
****Do want to have or avoid Homopolymer runs? Why? And what are they anyways?
Homopolymer run = 4+ identical bases in a row
AVOID
Can increase chances of False priming sites
Can cause slippage in annealing of primers
*What does it mean by slippage in annealing?

*****Why do we want a GC rich end at the 3’. What is the term for this 3’ end?
GC clamp or ANCHOR = allows primer to have complete primer binding due to stronger binding of GC
Increases specificity + accuracy
Increases stringency
****What are the 2 types of complementary sequences we want to AVOID with primers?
Between primers
Within primers
****What ISSUES do Complementary sequences between primers cause? what about within primers
Between = Primer Dimers
bp between 2 diff primers
Within = Primer Hairpins
Normally found with homopolymer runs
****How do Complementary sequences causing Primer Dimers cause issues for the rxn?
Primer dimers become COMPETITION for the template + remove available primers for the rxn
******Does the primer sequence have to match the target DNA exactly?
NO
except at 3’ end
*****Why does the Primer sequence have to match the target DNA exactly at the 3’ end but not anywhere else?
Stringency/Specificity depends SOLEY on where 3’-OH binds as that where polymerization begins
5’ end doesn’t contribute to polymerization and only a little to specificity
*****Since the 5’ end doesn’t have to match the target DNA. What can you do using it?
ADD EXTRA SEQUENCES useful for downstream applications aka future experiments (eg. Restriction enzyme site)
Although it is not complementary to the template, in LATER CYCLES the added extra sequence will be replicated as well
****Does the 5’ end matter more or less as the primer sequences gets longer
Less
contributes less and less to specificity as sequence gets longer
What programs are used for primer design? Are the primers given always exactly what you need?
BLAST + Primer 3
No primers given are sometimes wrong
****What 4 things do you need to analyze the given primers for?
Primer-dimer possibility
Other locations on template genome that give false priming sites
Similar Tm between primers
Perfect base pairing at 3’ end
*****What concentration of PRIMER should be used + why?
0.2 micromolar
Too low = not enough product
Too high = get non specific products
WHICH step of the cycle should be ADJUSTED relative to the primers??
Match ANNEALING temp to primers
the step where the primers attach/anneal to DNA
****What happens if the Annealing temp is too low?WHY?
Get low specificity + False priming
DNA likes to be double stranded therefore will reanneal to itself instead to primers
What is Taq
The main polymerase of PCR
Where is Taq isolated from?
Thermus Aquaticus bacterium
What is the optimum temp for Taq?
72 degrees Celsius
******What is an ISSUE with Taq?
Has NO PROOF READING = error prone
****What is the error rate of Taq?
1 in 10 000 nucleotides = mismatch occurs
****Why is Taq so error prone?
It lacks the 3’-5’ EXONUCLEASE activity = cannot correct mismatches
*****When is Taq best used? Why?
More error prone thus best used in: PCR when Sequence fidelity/exactness = less important
also CHEAP
*****What is ONE PRO of Taq despite its relatively high error rates?
It’s PROCESSIVE
*****Define PROCESSIVE and why it is a good thing.
Ability for (DNA POLY) to add nucleotide without frequently dissociating from template strand.
allows for RAPID + EFFICIENT synthesis of long stretches of DNA
also reduces errors of rebinding RNA poly to template
******Is Taq the only possible polymerase for PCR?
NO
What are the other 2 possible polymerases talked about in class for PCR?
Pfu
Phusion
What organism is Pfu Isolated from?
Pyrococcus Furiosus
*****What ADVANTAGE does Pfu have over Taq?
Has 3’-5’ Exonuclease activity = Much LOWER ERROR rate than Taq
******Does Pfu have any DISADVANTAGES?
YES
its LESS PROCESSIVE than Taq
Dissociates much more often
***What is the rate of polymerization for Pfu?
0.5 kb/min
*****Why does Pfu being less processive matter? Can’t you just increase extension times?
Heat degrades Stuff
Also with it falling off so often, it means it also needs to anneal correctly more times for an accurate replication
If both Taq + Pfu are not ideal What is the solution?
PHUSION
Taq/Pfu mix
****What is Phusion?
Fusion of two different enzymes with different activities required for DNA polymerase activity
*****What is Phusion normally a Fusion of?
DNA binding domain of a PROCESSIVE DNA poly (eg. Taq) + Polymerization Domain (pfu)
What advantages does Phu have over Taq + Pfu
Faster than pfu (processivity enhancing domain)
Higher fidelity than Taq (3’-5’ exonuclease activity = proof reading)
Why not use phu all the time?
Expensive
*****When is it Important to use Phu? (2 scenarios) WHy?
Expression CLONING
SEQUENCING
Accuracy of sequence = very important for both
OPTIMIZING PCR: What does a good Electrophoresis look like and what does a bad one look like?
Good = single amplified product
Bad = Smear +/or Multiple products
What is the most likely reason for a bad PCR?
YOU
forgot to add something
Just do it again the exact same way before trouble shooting
****Is a Smear above or below the Desired PCR worse?
Above
normally due to the 2 reasons listed above
****What is a smear on the BOTTOM normally due to?
Primer Dimers which is ok in small amounts
***What causes the primer dimers that show up as a smear on the Bottom?
Too high of a [Primer]
*****What are the 6 causes of Bad PCR results?
Annealing temp too low or Mismatched Primer Tm’s
Too much target DNA (the sequence meant to be amplified)
Too many cycles
Too much Mg2+
Non-optimal cycle parameters
Ramp time
****What is the outcome seen with Annealing temps too low? Why?
Multiple bands
Low temps = Low specificity = mis/false priming
****What are THREE WAYS to optimize Issues related to too low Annealing temps?
adding DMSO
Gradient PCR
Touchdown PCR
*****How does adding DMSO optimize Annealing temp?
Mimics base paring + Stabilizes Double Helix = can increases annealing temp slightly
Pairs with DNA template to prevent reannealing + allows Primers to bind more accurately with their complement
INCREASE SPECIFICITY
*****What is gradient PCR?
Thermocycler that allows different conditions (annealing temp) to occur in each row or column
Can run a number of different annealing temps in a single PCR run
*****How does adding Gradient PCR optimize Annealing temp?
Will show the optimal annealing temp for that rxn
****What does Touchdown PCR do? (2 things)
Decrease off-target priming
Increase specificity
*****How does adding Touchdown PCR optimize Annealing temp? What is the mechanism behind it?
Create conditions of High stringency in EARLY PCR cycle
****How are Conditions of High stringency created in the early cycles with Touchdown PCR?
Use a HIGHER than predicted ANNEALING TEMP
+ 10
Temp is gradually decreased in future cycles until it meets the typical parameters of Primers
****What does the High Annealing temps in the early stages of touchdown PCR Ensure?
Ensures Dominant product is amplified in all later cycles
by increasing specificity + decreasing false priming
****Why would adding too much target DNA cause bad PCR results?
All that is added initially in the tube present in the tube and run on the gel = multiple bands
****What is the common outcome of having too many CYCLES of PCR?
Very common source of SMEARS + Multiple bands
****Around how many replications do you start to see off target effects?
60 cycles
72 = giant smear
****Why would adding too much Mg2+ cause bad PCR results?
HIGH Mg2+ ==> Increases stability of the double helix + increases the tolerance for base mismatch
Also Mg2+ binds the dNTPs for DNA synth
****Why does LOW Mg2+ increase specificity?
Low Mg2+ decreases stability of the Hydration shell
Therefore = Requires HIGHER bp SPECIFICITY to anneal (harder to base pair unless its perfect)
What is the ideal Mg2+ concentration?
1-2.5 mM
*****Non-optimal parameters: Primer Extension time What happens when it’s too long or too short? What do you see on the gel?
Long = SMEAR
non specific primer. Gives more chances for primers to false prime
Short = No products
****What is the standard Extension rate for Taq, Pfu + Phusion?
Taq = 1 kb/min
Pfu = 0.5 kb/min
Phusion = 2kb/min