Chapter 9
1/56
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
57 Terms
Friedrich Miescher
the first chemist to isolate DNA
studied pus samples obtained from hospital bandages at Tubingen University (Germany)
separated the nuclei from the cytoplasm of white blood cells, isolated “nuclein” in 1869
nuclein
the substance isolated from white blood cells by Friedrich Miescher
contained large amounts of phosphorous, but no sulfur
based on lack of sulfur, concluded: not a protein
known today as DNA (deoxyribonucleic acid)
Phoebus Levene
incorrectly proposed the tetranucleotide hypothesis
(made other contributions to genetics, but those aren’t important)
tetranucleoide hypothesis
proposed by Phoebus Levene
DNA is composed of repetitions of the same sequence of four nucleotides (ATGC)
now known to be incorrect
four genetic properties of unknown genetic material
proposed by Frederick Griffith:
capable of replication
capable of storing information
complex enough to generate thousands of gene products
any changes in genetic material (mutations) should directly lead to changes in phenotype (now known that this is not always true)
Frederick Griffith
proposed the four properties of unknown genetic material
conducted the Griffith experiments, demonstrated genetic transformation in S. pneumoniae
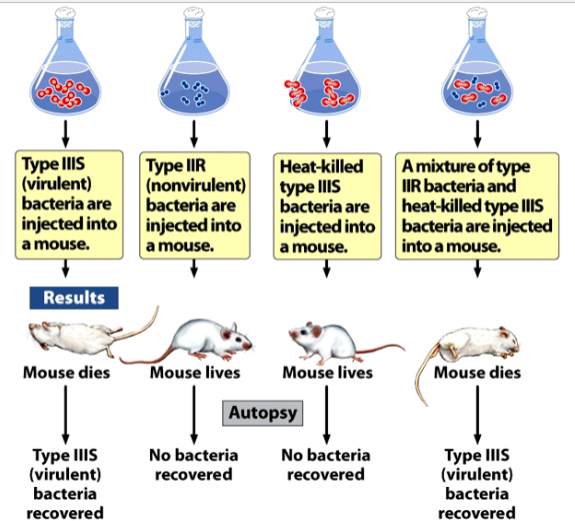
the Griffith experiments
demonstrated genetic transformation in bacteria Streptococcus pneumoniae
strain IIIS (smooth) is virulent; causes fatal pneumonia in mice. Cells can be recovered from lungs of killed mice
strain IIR (rough) is nonvirulent; mice injected with it survive, not cells can be recovered from these mice.
Heat kills IIIS cells, but an unknown “transforming principle” survives and is capable of transforming IIR cells into IIIS cells
results:
mice injected with IIIS die, IIIS bacteria are recovered
mice injected with IIR survive, no bacteria are recovered
mice injected with heat-killed IIIS survive, no bacteria are recovered
mice injected with a mixture of IIIR and heat-killed IIS die, IIIS bacteria are recovered
transformation
a heritable change in a cell or organism brought about by exogenous DNA
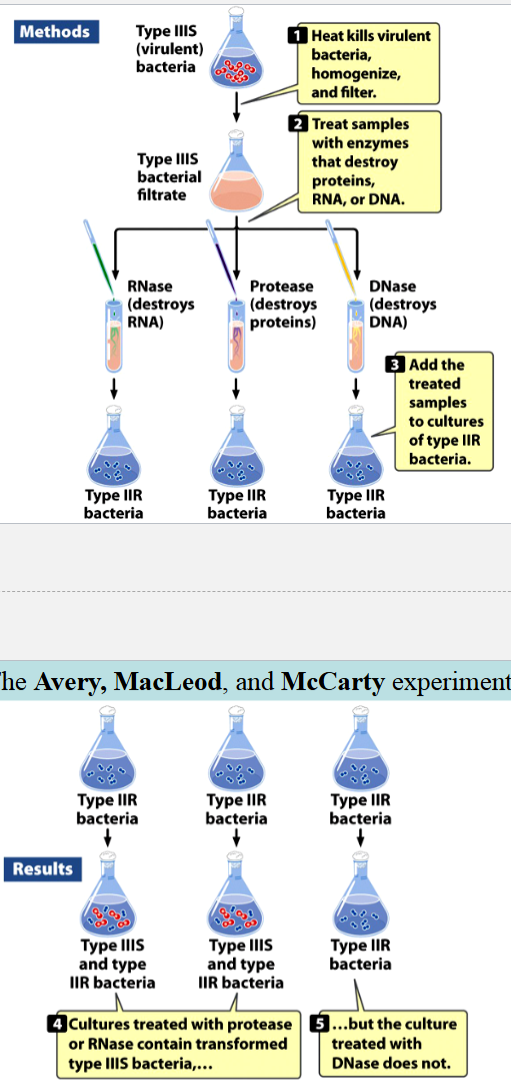
the Avery, MacLeod, and McCarty experiments
identified Griffith’s transforming principle
IIIS cultures (in liquid media) were centrifuged, collected, heat-killed, lysed, and homogenized
enzymes and extractions eliminated with carbohydrates, lipids, and proteins
resulting solution had nitrogen:phosphorus ratio consistent with a nucleic acid, and was still capable of transforming IIR into IIIS cells
either proteins, RNA, or DNA were destroyed with enzymes
resulting mixtures were added to cultures of IIR cells
cultures with RNA and proteins destroyed contained transformed IIIS, culture without DNA did not
conclusion: IIIS DNA can transform IIR → IIIS cells
protease
enzyme that destroys proteins
used in the Avery, MacLeod, and McCarty experiments
ribonuclease (RNase)
enzyme that destroys RNA
used in the Avery, MacLeod, and McCarty experiments
deoxyribonuclease (DNase)
enzyme that destroys DNA
used in the Avery, MacLeod, and McCarty experiments
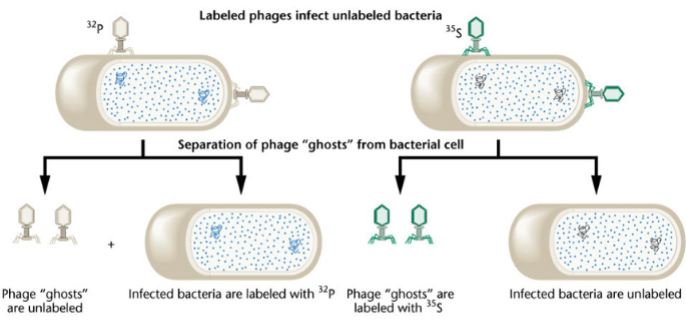
the Hershey & Chase experiments (1952)
showed that DNA is the infectious agent in bacteriophages
1) infect bacteria in radioactive media with T2 phages
2) infect cells in non-radioactive media with radioactive phages
3) after infection, separate phages from cells in a blender, centrifuge the suspension
32P phages have radioactive DNA, 35S phages have radioactive proteins
bacteria infected by 32P phages incorporate radioisotope 32P; radioactivity will be found in cells at the bottom of the centrifuge tube (the pellet)
bacteria infected by 35S phages, the radioisotope stays outside of the cells; radioactivity will be found in the supernatant
conclusion: DNA, not protein, is the genetic material in phages
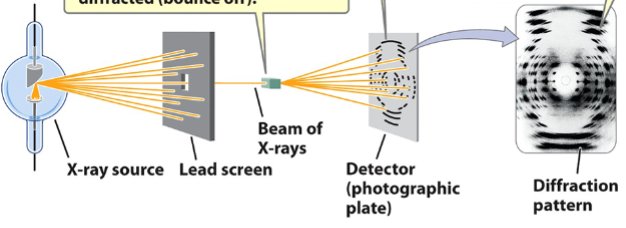
X-ray diffraction
used to elucidate the structures of molecules
steps:
crystals of substance are bombarded with X-rays through a lead screen. The X-rays get diffracted (bounce off of) the substance
the spacing of the atoms within the sample crystal determines the diffraction pattern. Diffraction pattern appears as spots on a photographic film
the diffraction pattern provides information about the structure of the molecule
Rosalind Franklin
credited with elucidating the helical structure of DNA
carried out X-ray crystallography of DNA fibers in 1952
photograph 51
diffraction profile was consistent with helical structure, possible double helix, with a sugar-phosphate backbone outside
worked in Maurice Wilkins’ lab at King’s College, London

Erwin Chargaff
studied the base-composition of DNA
formed Chargaff’s rules
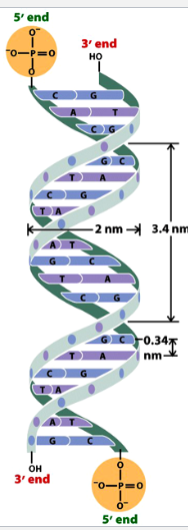
the Watson-Crick model
DNA (deoxyribonucleic acid) is a double helix with antiparallel strands
strands are made up of a sugar + phosphate backbone and nitrogenous bases paired at the center by hydrogen bonds
alternating major and minor grooves
the length of one whole turn of the helix is ~10 base pairs (bp)
James Watson and Francis Crick “proposed” the double-helix model of DNA, published model in 1953, were awarded the Nobel Prize in physiology or medicine in 1962
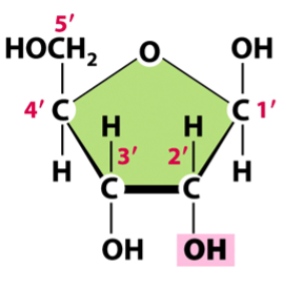
ribose
a pentose sugar present in RNA, makes up the backbone along with phosphate
difference from deoxyribose: has an OH group at 2’C instead of a second H
deoxyribose
a pentose sugar present in DNA, makes up the backbone along with phosphate
difference from ribose: lacks an OH at 2’C, instead has a second H (only 2 OH groups directly connected to the pentose (exclude the OH connected to 5’C))
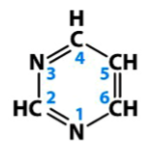
purines
one group of nitrogenous bases in nucleic acids
double carbon-nitrogen aromatic structures
in nucleic acids, include adenine and guanine
basic structure is depicted
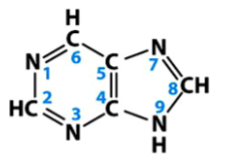
pyrimidines
one group of nitrogenous bases in nucleic acids
single carbon-nitrogen aromatic ring structures
in nucleic acids, include cytosine, thymine, and uracil
basic structure is depicted
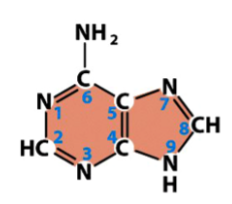
adenine (A)
a purine
found in both DNA and RNA
pairs with thymine in DNA
pairs with uracil in RNA
from basic purine structure, only difference is an added amine (NH2) at one C
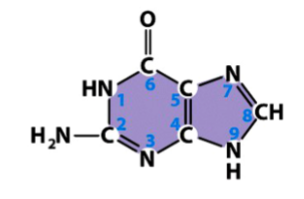
guanine (G)
a purine
found in both DNA and RNA
pairs with cytosine in both DNA and RNA
from basic purine structure, a carbonyl (double bonded oxygen to carbon) is added to one C, the adjacent double bond is removed, an amine (NH2) is added to another C, and add Hs to Ns missing a double bond
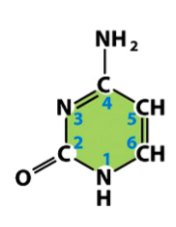
cytosine (C)
a pyrimidine
present in both DNA and RNA
pairs with guanine in both DNA and RNA
from basic pyrimidine structure, add an amine (NH2) to one C, add a carbonyl (O double bonded to C) to another C, remove the adjacent double bond, and add Hs to Ns missing a double bond
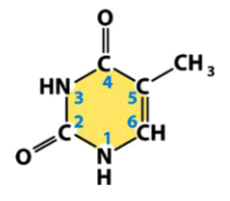
thymine (T)
a pyrimidine
found only in DNA
pairs with adenine in DNA
from basic pyrimidine structure, add 2 carbonyls (O double bonded to C), remove the double bonds adjacent to both, add a methyl (CH3) to another C, and add Hs to Ns missing double bonds
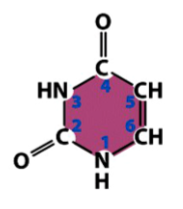
uracil (U)
a pyrimidine
found only in RNA
pairs with adenine in RNA
from basic pyrimidine structure, add carbonyls (O double bonded to C) to 2 Cs, remove the adjacent double bonds, and add Hs to Ns missing a double bond
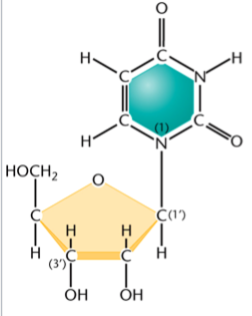
nucleoside
a unit made of a sugar and nitrogenous base
no phosphate
(think: no T in the name, no phosphaTe)
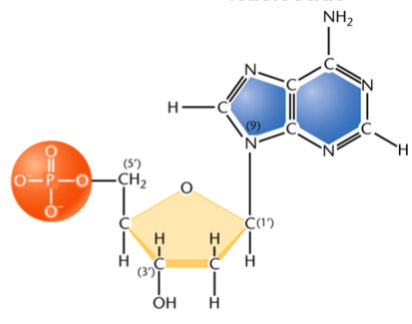
nomenclature of ribonucleosides
nucleotide (root) + either -sine or -dine (suffix)
suffix is -sine for purines, -dine for pyrimidines (both have a d in the name)
e.g. adenosine, cytidine, guanosine, uridine
nomenclature of deoxyribonucleosides
deoxy- (prefix) + nucleotide (root) + either -sine or -dine (suffix)
suffix is -sine for purines, -dine for pyrimidines (both have a d in the name)
e.g. deoxyadenosine, deoxycytidine, deoxyguanosine, deoxyuridine
nucleotide
a unit made of a sugar, nitrogenous base, and phosphate(s)
(think: has a T in the name, has phosphaTe)
consist of ribonucleotides and deoxyribonucleotides
nomenclature of nucleotides
same as nomenclature of nucleosides, then add mono/di/tri/etc-phosphate
e.g. uridine monophosphate (UMP), cytidine diphosphate (CDP), deoxythymidine diphosphate (dTDP), adenosine triphosphate (ATP)
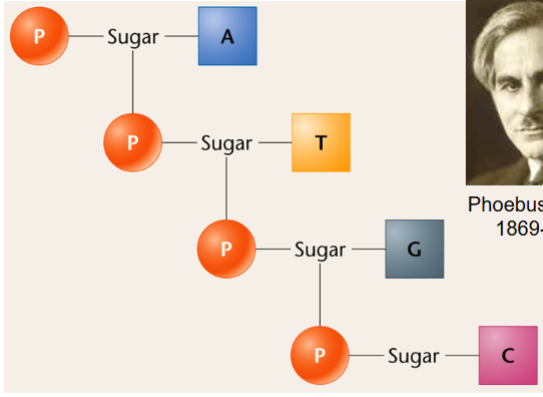
phosphodiester bond
joins the 3’ C of one nucleotide and the 5’ C of the next through 2 ester linkages
has oxygens (OR groups) on both sides
link nucleotides in the polynucleotide chains of DNA and RNA
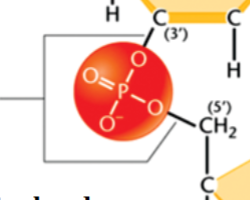
Watson-Crick base pairs
A-T and A-U: 2 hydrogen bonds form between the nucleotides
G-C: 3 hydrogen bonds form between the nucleotides
Chargaff’s rules
1) the proportion of A = proportion of T, and the proportion of G = proportion of C
2) there is an equal proportion of purines (A and G) and pyrimidines (C and T)
3) the proportion of C + G does not necessarily equal that of A + T
intramolecular hydrogen bonds
hydrogen bonds within a molecule
can form in most RNAs, e.g. transfer RNA (tRNA, because it is single-stranded)
results in endless possibilities for 3D structures
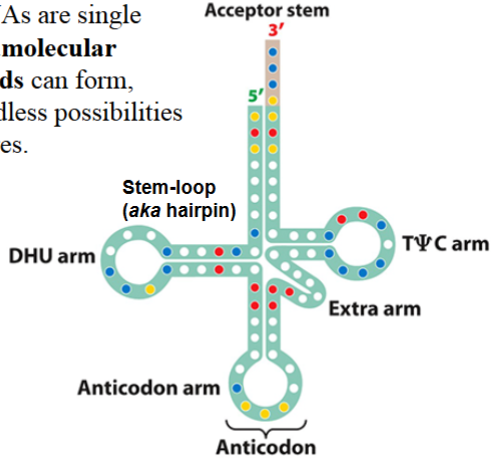
organization of DNA in eukaryotes
DNA is contained in nucleus as chromosomes
always present, but only visible (condensed chromosomes) during mitosis
organized in nucleosomes
adjacent nucleosomes pack together, form 30-nm chromatin fibers
higher order levels of organization results in the condensation of DNA into the visible chromosome beginning at prophase
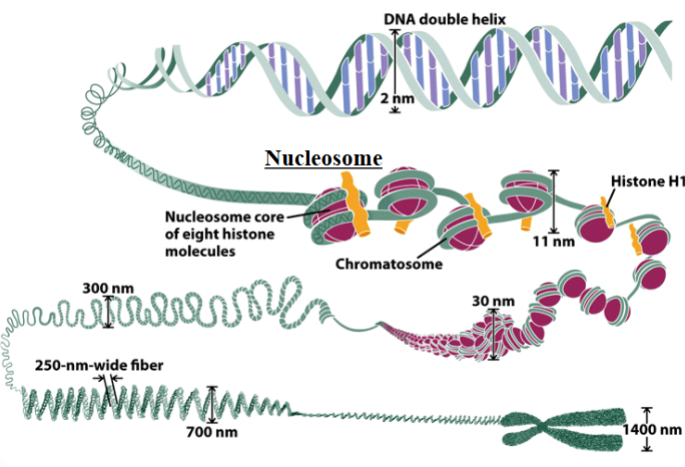
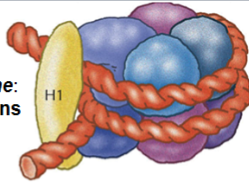
nucleosome
the basic unit of organization of DNA in eukaryotic nuclei
each unit consists of 2 turns of DNA (147 bp) wrapped around 8 histone proteins and held together by a ninth histone (histone 1)
isolation and purification of DNA
procedure for isolation is similar to that used by Avery, MacLeod, and McCarty
purification: precipitation of DNA by the addition of ethanol, then the redissolving of the precipitate
done in preparation for enzymatic manipulation
nucleases
enzymes that break up polynucleotide chains
can be exonucleases or endonucleases
exonucleases
enzymes that remove nucleotides one by one from the ends of a polynucleotide chain
endonucleases
enzymes that cleave phosphodiester bonds within a polynucleotide chain
can produce sticky ends or blunt ends
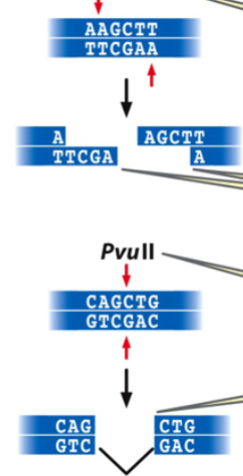
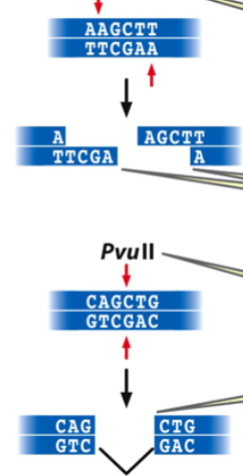
restriction endonuclease
an enzyme that recognizes a specific base pair sequence in DNA and makes a cut at that site
the bp sequence is a restriction site with a palindrome sequence
restriction digestion of DNA
results in restriction fragments: small linear fragments with either complementary single-strand sticky ends (cohesive tails) or blunt ends
type of end depends on the restriction enzyme
e.g. HindIII makes sticky ends, PvuII makes blunt ends
e.g. can manipulate plasmids
cloning vector
e.g. pUC18
has a polylinker region that contains restriction sites for several restriction endonucleases
any foreign DNA introduced in this region of the plasmid will be replicated every time the cell divides
ways to introduce DNA into cells
electric shock
heat shock
microinjection
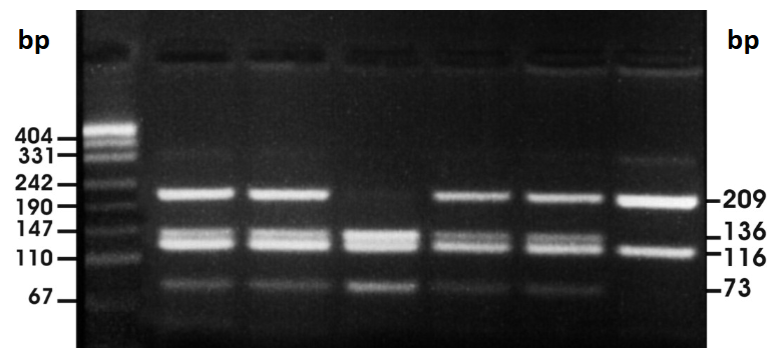
electrophoresis
a method for separating fragments of nucleic acids according to size
molecules migrate to agarose gel medium in electric field
because phosphate groups have negative charges, molecules migrate toward the positive end of the field
smaller molecules migrate faster (will end up closer to the positive end)
for nucleic acids, a dye is added to the gel. DNA fragments will appear as bands in the gel
preparation of an agarose gel
agarose gel: semisolid slab that has wells near one end for loading samples of nucleic acid
a tray with the gel is placed in an electrophoresis chamber, which is filled with buffer solution to cover the gel
DNA samples are loaded into the wells
electric field is applied; cathode (-) is placed at the sample end, anode (+) at the opposite end
ethidium bromide staining
used for electrophoresis of nucleic acids
fluorescent dye that becomes nested between the nitrogenous bases
can be excited by UV light to show bands of migrated DNA
presentation of DNA sequences
the sequence of nucleotides of only one strand is shown
e.g. present only the 5’ to 3’ strand, not the 3’ to 5’ strand as well
(the other strand is complementary, so it would just be the presented strand in backwards order)
conclusion of the Avery, MacLeod, and McCarty experiments
because only DNase destroyed the transforming substance, the transforming substance if DNA
the infectious agent in phages is
DNA
High G-C content DNA have higher
density and melting point
nucleoside
sugar + nitrogenous base
nucleotide
sugar + nitrogenous base + phosphate(s)
deoxyribose lacks the
2’ OH group
how do you number the carbons in the pentose sugars
1’ to 5’, right to left
what is the nucleosome core particle
eight histone proteins