tutorial 8: Barcoding presentation and ISOLATION, CLONING AND TRANSFORMATION OF PLASMID DNA Day 3 plasmid restriction and ligation
1/13
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
14 Terms
What makes a suitable barcode
Contain significant species-level genetic variation and divergence
Possess conserved flanking sites for developing universal PCR primers
for wide taxonomic applicationHave a short sequence length to facilitate current capabilities of DNA
extraction and amplification
Cytochrome c oxidase I (COX1)
Last enzyme in the respiratory electron transport chain in mitochondria
Found in nearly all eukaryotes and in multiple copies within an individual
More than 2% sequence divergence typical for closely related species but generally conserved within species
what is barcoding used for?
Taxonomy and phylogenetics
Identifying unknown species
Detecting invasive species
Species surveillance and conservation (monitor for signs of human interference, climate change, pollution, etc.)
Wildlife crime (products from endangered species or illegally
traded wildlife, poaching, etc.)Quality assurance and control (e.g., fish fraud)
Adulteration – test for presence of adulterants (e.g., mixed meats or other organisms presented as one)
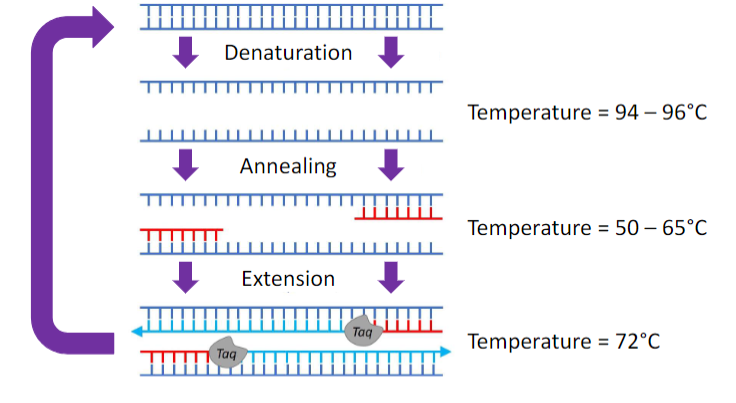
name the steps of PCR and the temperature range for each
see picture
purpose of Restriction endonuclease digestion of pUC18 and pKAN
pUC18: to open up the multiple cloning site (MCS) so that a fragment from
pKAN (containing the kanamycin resistance gene) can be inserted into the MCS
purpose of ligation reaction
ligases use ATP to link the phosphate group on the 5’ end of one DNA strand to the hydroxyl group on the 3’ end of the DNA strand through the formation of a phosphodiester bond
what are the possible products of a digestion of pUC18 with HinDIII and BamHI?
A fragment with lacZ and rep for replication, with sticky end for HinDIII and BamHI
A short fragment produced from the MCS, with BamHI and HinDIII sticky ends
in the pUC18 plasmid, what is the role of LacZ in screening? what is the role of rep?
The LacZ polypeptide corresponding to wild type beta-galactosidase and is essential for blue/white screening.
The indicated rep region is sufficient to promote replication
what is the replicon made of?
The replicon is comprised of the origin of replication (ORI) and all of its control elements. The ORI is the place where DNA replication begins, enabling a plasmid to reproduce itself as it must to survive within cells.
describe the E. coli lac operon region on the pUC18
the region of E.coli lac operon containing a CAP protein* binding site, promoter Plac, lac repressor binding site and the 5’-terminal part of the lacZ gene encoding the N-terminal fragment of beta-galactosidase.
This fragment is capable of intra-allelic (alpha) complementation with a defective form of beta-galactosidase encoded by the host.
Bacteria that can synthesize both fragments of the enzyme form blue colonies on media with X-Gal.
Insertion of DNA into the MCS located within the lacZ gene (codons 6-7 of lacZ are replaced by MCS) inactivates the N-terminal fragment of beta-galactosidase and abolishes alpha- complementation.
Bacteria carrying recombinant plasmids therefore give rise to white colonies
what is CAP?
The catabolite activator protein (CAP) is a transcriptional activator that must bind to cAMP to activate transcription of the lac operon by RNA polymerase. cAMP is a derivative of adenosine triphosphate (ATP) and used for intracellular signal transduction in many different organisms
what are the two possible products of Digestion of pKAN with HinDIII and BamHI
A fragment with kanamycin resistance gene, but no origin of replication, with sticky ends for BamHI and HinDIII
A fragment with origin of replication but not kanamycin resistance, with sticky ends for BamHI and HinDIII
why are buffers containing low EDTA concentrations often used?
Buffers containing low concentrations of EDTA (1mM) are often used to protect DNA from nuclease degradation during storage, but EDTA can interfere with restriction enzyme digestion if the final concentration in the reaction is high
describe ligation reactions
A group of enzymes called DNA ligases seal cut or broken DNA by creating a covalent linkage between the 5' phosphate group of one chain with the adjacent 3' -OH group of another