👎 Week 11 - Mutations
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
Mutations and DNA Repair: Section Outline
Mutation – the source of all genetic variability
Types of gene mutations
Functional effects of mutations
Suppressor mutations
Mutation rates
Molecular mechanisms of mutation
Mechanisms of DNA repair
Learning Objectives: Mutations and DNA Repair
Explain why DNA mutations are the source of phenotypic variation
Explain why many DNA mutations do not affect phenotype
List three ways DNA mutations can occur
Identify how mutations are detected and repaired
Recognize that mutations often pass through an intermediate stage that can be repaired to avoid mutation
Describe the types of chromosomal rearrangements
Mutations
Definition
Change in the nucleotide sequence of an organism’s genome
Source of all genetic variability
Causes
Agents that damage DNA: UV light, certain chemicals, viruses
Random errors during DNA replication
Effects on Phenotype
Generate nonfunctional proteins
Alter protein function
Change when and where a gene is expressed
Mutant
Organism carrying one or more mutations in its genetic material
Mutations
Occurrence
Spontaneous – occur naturally at low rates
Induced – caused by mutagens
Effects
Change DNA sequence
May alter or eliminate encoded proteins
Phenotypic consequences may or may not occur
Genetic Variation and Mutation
Mutation
Occurs in all organisms with genetic material, from viruses to humans
Source of all genetic variation
Natural Selection
Preserves gene combinations that are best adapted to the environment
Drives evolution
Recombination During Meiosis
Homologous chromosomes exchange segments
Rearranges genetic variability into new gene combinations
Key Point
Mutation is the fundamental source of all genetic variability
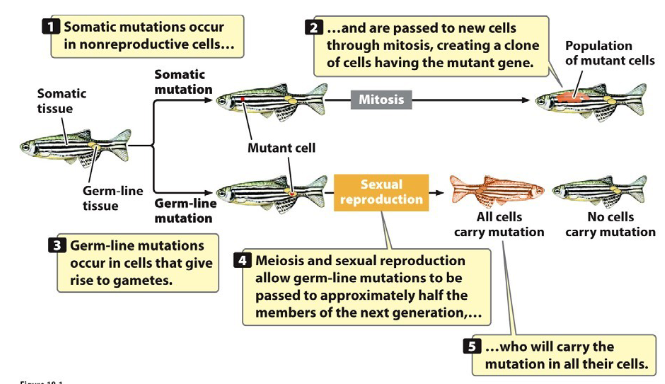
Types of Mutations
Somatic Mutations
Occur in somatic (body) cells
Affect only the descendants of that cell
Not transmitted to offspring
Germinal (Germ Line) Mutations
Occur in germ-line (reproductive) cells
Transmitted to progeny through gametes
Types of Mutations
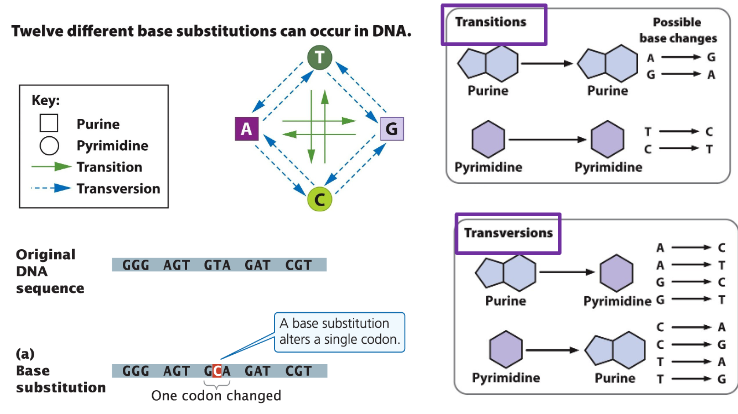
A. Base Substitutions
Transition
Replacement of a purine with another purine (A ↔ G) or a pyrimidine with another pyrimidine (C ↔ T)
Transversion
Replacement of a purine with a pyrimidine or vice versa
B. Insertions and Deletions
Frameshift Mutations
Addition or loss of nucleotides that shifts the reading frame
In-Frame Insertions and Deletions
Addition or loss of nucleotides in multiples of three, preserving the reading frame
C. Tautomeric Shifts
Movement of hydrogen atoms within bases
Can cause mispairing during DNA replication
Base Substitutions - 12 Possibilities
Transition (same base type switch)
Replaces a pyrimidine with another pyrimidine (C ↔ T) or a purine with another purine (A ↔ G)
Transversion (opposite base type switch)
Replaces a pyrimidine with a purine or a purine with a pyrimidine
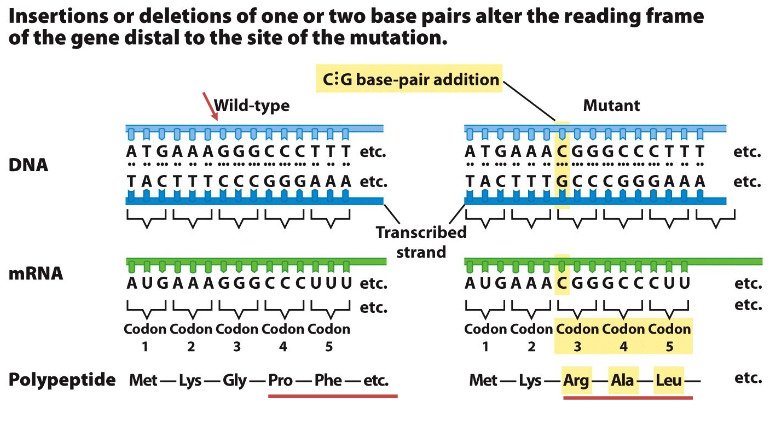
Frameshift Mutations
Definition
Insertion or deletion of one or two base pairs that changes the reading frame of the gene
Effect on Protein
Protein sequence changes dramatically after the site of the mutation
Tautomeric Shifts
Definition
Reversible change in the location of a hydrogen atom in a molecule
Alters the molecule from one isomer to another
Specifically, movement of H⁺ atoms within purine or pyrimidine bases
Occurrence
Rare and can happen spontaneously during DNA replication
Alters base pairing and can cause spontaneous mutations
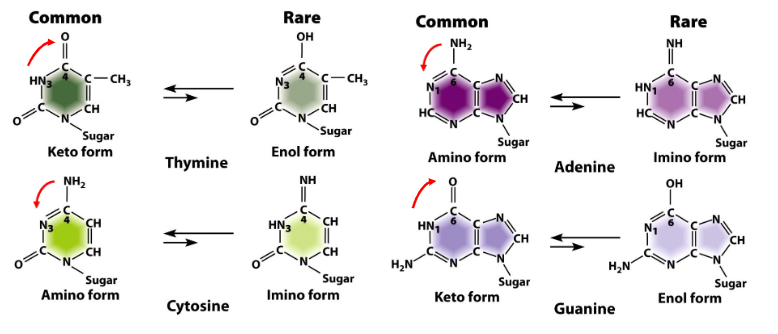
Tautomeric Shifts
Effect on Base Pairing
Can generate rare A:C and G:T base pairs during DNA replication
Mechanism
Occurs when bases adopt their rare enol or imino forms
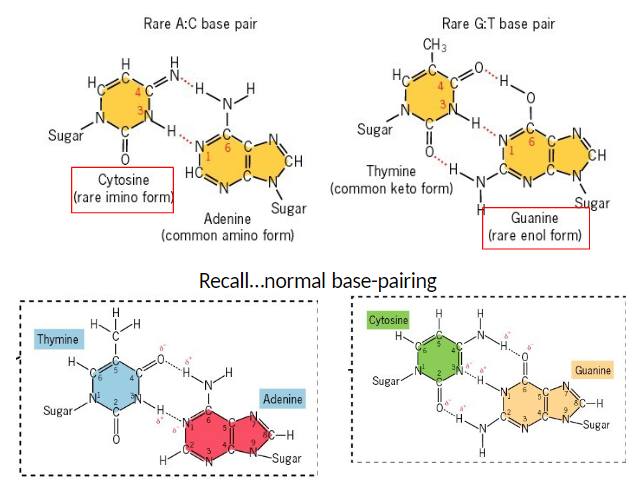
Tautomeric Shift Mutations: Example Mechanism
Normal Base
Guanine (G) in its normal keto form pairs correctly with cytosine (C)
Rare Tautomeric Form
Guanine shifts to its rare enol form (G*)
This form mispairs with thymine (T) during DNA replication
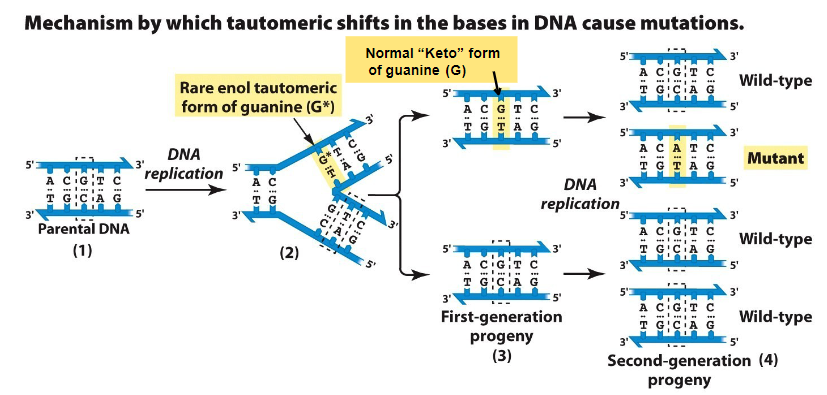
Replication Process
Parental DNA contains G in normal keto form
During replication, G* pairs with T instead of C
First-generation progeny contains half normal half G paired with T
Second-generation contains ¾ normal and ¼ mutant since G paired with C correctly from first-gen progeny parent but T paired with A which wasn’t there before
Result
A single base change occurs: G → A transition mutation
Mutation appeared in second gen progeny
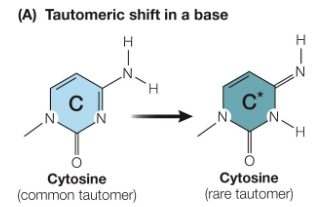
Tautomeric Shifts
Definition
Bases have two isomers called tautomers
When a base temporarily adopts its rare tautomeric form, it can pair with the wrong base
This leads to a mismatch during DNA replication
Types of Point Mutations
Missense Mutation
Codon change results in a different amino acid
Nonsense Mutation
Codon change converts a sense codon into a stop codon
Silent Mutation
Codon change results in a synonymous codon, amino acid remains the same
Functional Effects of Mutations
Missense Mutation
Base substitution changes an amino acid in the protein
Nonsense Mutation
Base substitution converts a sense codon into a stop codon (UAG, UGA, UAA)
Silent Mutation
Base substitution changes the codon but still specifies the same amino acid
Key Point
Mutations are classified based on how they affect gene or protein function

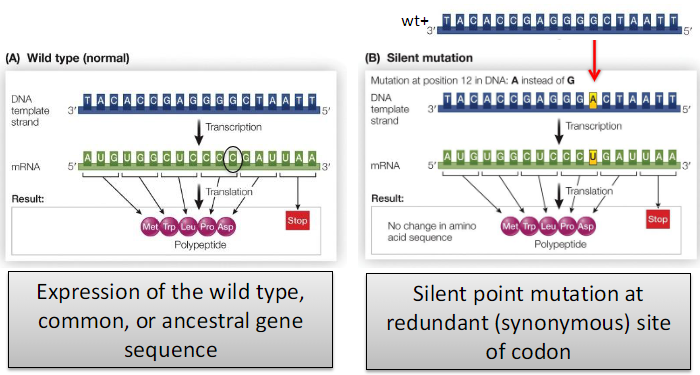
Silent Point Mutation
Definition
Result from the gain, loss, or substitution of a single DNA base pair
Effect in Coding Regions
May be silent or may change the amino acid sequence of the protein
Silent Point Mutation
Occurs at a redundant (synonymous) site of a codon
Expression of the wild-type gene sequence is NOT maintained
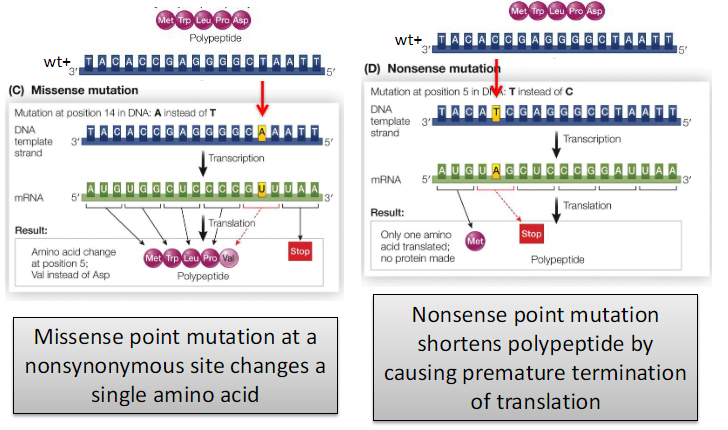
Missense Point Mutation
Effects on Protein
May change a single amino acid (missense)
May cause loss of amino acids at the carboxyl terminus
Missense Point Mutation
Occurs at a nonsynonymous site of a codon
Changes the identity of a single amino acid
Amino acid properties can change (e.g., negatively charged → nonpolar/hydrophobic)

Missense vs. Nonsense Mutations
Missense Point Mutation
Occurs at a nonsynonymous site of a codon
Changes a single amino acid in the protein
Nonsense Point Mutation
Converts a sense codon into a stop codon
Causes premature termination of translation
Shortens the polypeptide
General Effect
Point mutations may change an amino acid or cause loss of amino acids at the carboxyl terminus of the protein
Point Mutations in Coding Regions
Frame-Shift Point Mutation
Insertion or deletion changes the reading frame
Drastically alters the sequence of the resulting polypeptide
Loss-of-Stop Mutation
Eliminates the normal stop codon
Translation continues until a new stop codon is reached
Can extend or alter the C-terminal end of the protein

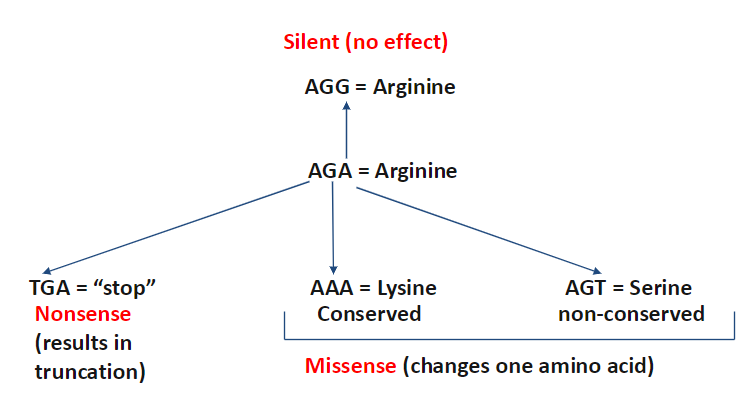
Effects of Single Base Mutations
Silent Mutation
No effect on protein sequence
Example: AGG → Arginine, AGA → Arginine
Nonsense Mutation
Changes a codon to a stop codon
Results in truncation of the protein
Example: TGA = stop
Missense Mutation
Changes one amino acid in the protein
Can be conserved (similar properties, e.g., AAA → Lysine)
Or non-conserved (different properties, e.g., AGT → Serine)
Types and Effects of Gene Mutations
Base Substitutions (Point Mutations)
Can be transitions or transversions
Silent Mutation – Codon changes but still specifies the same amino acid, no effect on protein
Missense Mutation – Codon changes to a different amino acid
Can be conserved (similar properties, e.g. AAA → Lysine)
Or non-conserved (different properties, e.g. AGT → Serine)
Nonsense Mutation – Codon changes to a stop codon (UAG, UGA, UAA)
Causes premature termination, truncating the polypeptide
Frameshift Mutations
Insertion or deletion of one or two nucleotides
Alters the reading frame
Drastically changes amino acid sequence downstream
Loss-of-Stop Mutation
Eliminates normal stop codon
Translation continues to next stop codon, extending or altering the C-terminal end
Insertions and Deletions (In-Frame)
Addition or loss of nucleotides in multiples of three
Preserves reading frame
Adds or removes amino acids without shifting downstream sequence
Tautomeric Shifts
Bases temporarily adopt rare forms (enol or imino)
Mispairing occurs during DNA replication (e.g., A:C, G:T)
Can result in point mutations like transitions or transversions
A. Base Substitutions
Transition
Replacement of a purine with another purine (A ↔ G) or a pyrimidine with another pyrimidine (C ↔ T)
Transversion
Replacement of a purine with a pyrimidine or vice versa
B. Insertions and Deletions
Frameshift Mutations
Addition or loss of nucleotides that shifts the reading frame
In-Frame Insertions and Deletions
Addition or loss of nucleotides in multiples of three, preserving the reading frame
C. Tautomeric Shifts
Movement of hydrogen atoms within bases
Can cause mispairing during DNA replication
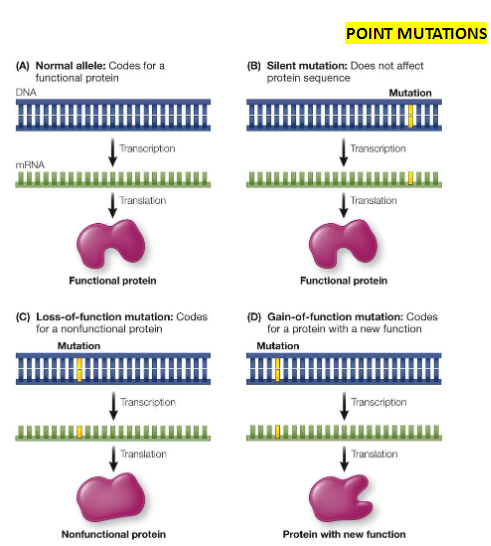
Functional Effects of Mutations
Silent Mutations
Do not affect protein function
Loss-of-Function Mutations
Prevent gene transcription or produce nonfunctional proteins
Usually recessive
Gain-of-Function Mutations
Produce a protein with altered or new function
Usually dominant
Common in cancer cells
Point Mutations
Forward Mutation – Wild-type sequence changes to mutant
Reverse Mutation – Mutant sequence changes back to wild-type
Types of Functional Mutations
Neutral Mutation
A missense mutation changes an amino acid to one with similar chemical properties (e.g., glycine → alanine)
No observable effect on protein function
Loss-of-Function Mutation
Mutations cause complete or partial loss of normal protein function
Example: cystic fibrosis caused by loss-of-function mutation in the CF gene
Gain-of-Function Mutation
Mutations produce a protein with a new or abnormal function
Example: mutation in a cell surface growth receptor that stimulates growth without the growth factor
Conditional Mutation
Mutation is expressed only under specific conditions
Example: temperature-sensitive mutation observable only at extreme temperatures
Gain-of-Function Mutation (GOF) *Don’t memorize name
Example
A transcription factor normally active only in stem cells for larval leg development is mistakenly expressed in stem cells for larval antenna development
Example gene: Antennapedia
Effect
Causes a new or abnormal function, such as leg structures forming where antennae should develop
Conditional Mutation
Temperature-Sensitive Allele
Functional only under certain temperatures
Example: Fruit fly with a mutation in a wing development gene
Protein is non-functional at low temperatures (cold-sensitive)
Protein is functional at warmer temperatures
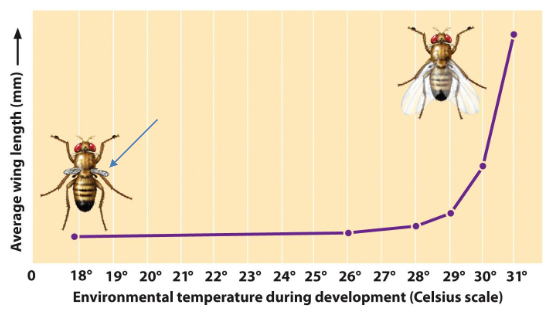
Conditional Mutations
Definition
Affect the phenotype only under specific environmental conditions
Wild-type phenotype is expressed under other conditions
Example
Coat color in some cats and rabbits
Temperature-sensitive mutant protein is inactive in warmer body regions → pale fur
Protein is active in cooler extremities (ears, nose, feet) → dark fur
Lethal Mutation
Definition
Mutation that causes premature cell death
Chromosomal Mutations
Definition
Extensive changes in genetic material involving long DNA sequences
Significance
Can provide genetic diversity for evolution by natural selection
Often deleterious
Mechanism
Chromosomal rearrangements involve double-strand breaks
Aberrant crossover between homologous or nonhomologous chromosomes can cause rearrangements
Radiation can induce double-strand breaks, and repair may join non-homologous ends
Effect on Cell
Multiple chromosome breaks can cause cell death due to loss of DNA fragments or triggering of apoptosis
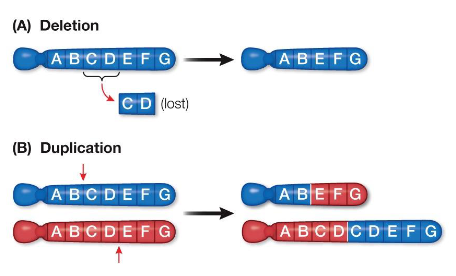
Chromosomal Rearrangements: Deletions and Duplications
Deletion
Loss of a chromosome segment
Can have severe or fatal consequences
Duplication
A portion of a chromosome is replicated, creating multiple copies
Occurs when homologous chromosomes break at different positions and reconnect incorrectly
One chromosome loses the segment, the other gains two copies
Extra copies can lead to overexpression of genes
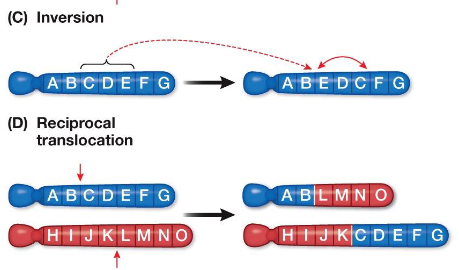
Chromosomal Rearrangements: Inversions and Translocations
Inversion
A chromosome segment breaks and rejoins in the reverse orientation (“flipped”)
Can result in loss-of-function mutations
Translocation
A chromosome segment breaks off and inserts into another non-homologous chromosome
Often involves reciprocal exchange of segments between chromosomes
Can place a gene next to a new control region, altering its expression
Forward and Reverse Mutations
Forward Mutation
Change from wild-type to mutant form
Reversion Mutation
A second mutation that restores the wild-type phenotype
True Reversion – Converts the mutant nucleotide sequence back to the original wild-type sequence
Suppressor Mutation
A second mutation that compensates for the effect of the original mutation without restoring the original nucleotide sequence
Intragenic Suppressor – Occurs within the same gene
Intergenic Suppressor – Occurs in a different gene
Original and Suppressor Mutations
Original Mutation
The initial change in the DNA sequence that causes a problem
Example: produces a nonfunctional protein
Suppressor Mutation
A second mutation elsewhere in the genome
Does not revert the original mutation
Compensates for the defect caused by the original mutation
Example: restores some protein function, helps the cell compensate, or alters interactions to reduce negative effects
Intragenic Suppressor Mutations
Definition
A second-site mutation that hides or suppresses the effect of the first mutation
Not a reverse mutation
Types
Intragenic Suppressor – Occurs within the same gene as the original mutation
Intergenic Suppressor – Occurs in a different gene (implied from earlier notes)
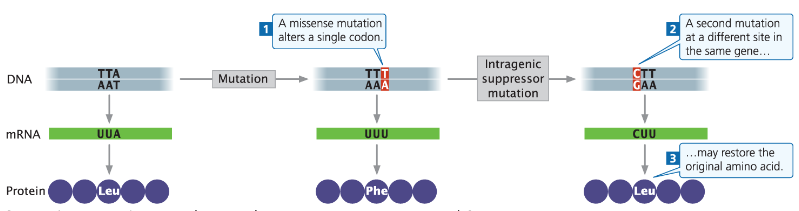
Intergenic Suppressor Mutations
Intergenic Suppressor
A suppressor mutation that occurs in a different gene from the original mutation
Mechanism
The second mutation can be a gain-of-function mutation
Example: a mutated tRNA acquires a novel function, such as binding to a stop codon to suppress a nonsense mutation
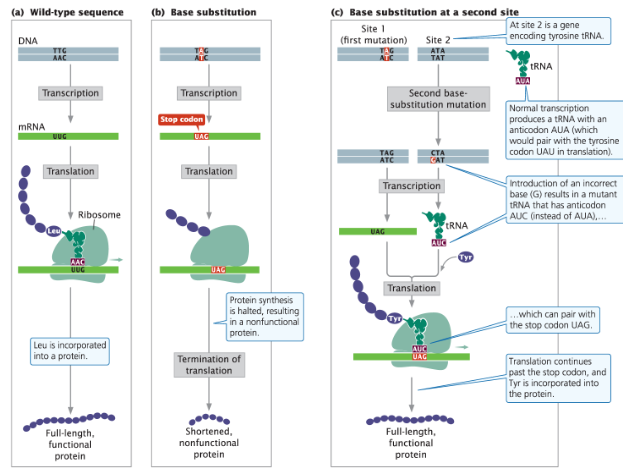
Relationship to Reverse and Suppressor Mutations
Forward Mutation
Changes the wild-type gene into a mutant phenotype
Reverse Mutation
Restores the wild-type gene and phenotype
Suppressor Mutation
Occurs at a different site than the original mutation
Individual carries both the original mutation and the suppressor mutation
Phenotype appears wild-type despite the original mutation
Example Genotype
Wild type: A⁺ B⁺
Forward mutation produces a mutant
Suppressor mutation restores the wild-type phenotype without changing the original mutant nucleotide sequence
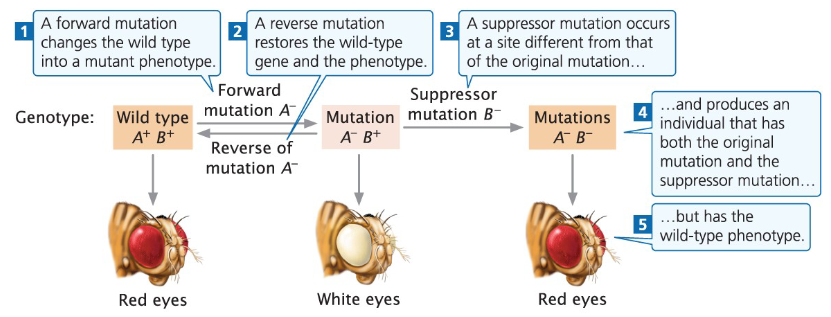
Example of Reversion and Suppressor Mutations
Harlequin Norway Maple Trees
Mutation affects chlorophyll production, resulting in partly albino leaves
Revertants
Occasionally, all-green leaves appear
Result of a secondary mutation that restores the wild-type phenotype
Masks the effect of the original mutation
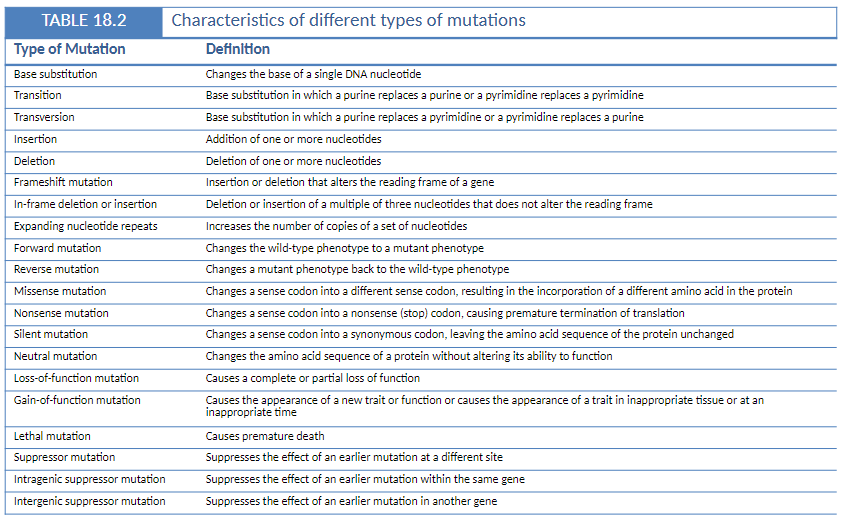
Summary of Mutation Types
Base Substitution
Change of a single DNA nucleotide
Transition
Purine ↔ purine or pyrimidine ↔ pyrimidine
Transversion
Purine ↔ pyrimidine
Insertion
Addition of one or more nucleotides
Deletion
Loss of one or more nucleotides
Frameshift Mutation
Insertion or deletion that alters the reading frame of a gene
In-Frame Deletion or Insertion
Deletion or insertion in multiples of three nucleotides
Does not change the reading frame
Expanding Nucleotide Repeats *NOT ON EXAM
Increase in the number of copies of a nucleotide sequence
Forward Mutation
Changes wild-type phenotype to mutant
Reverse Mutation
Restores mutant phenotype back to wild-type
Missense Mutation
Sense codon changes to a different sense codon
Incorporates a different amino acid
Nonsense Mutation
Sense codon changes to a stop codon
Causes premature translation termination
Silent Mutation
Codon changes but amino acid sequence remains unchanged
Neutral Mutation
Changes amino acid without affecting protein function
Loss-of-Function Mutation
Complete or partial loss of normal protein function
Gain-of-Function Mutation
New trait or function appears
Can be expressed at inappropriate tissue or time
Lethal Mutation
Causes premature death
Suppressor Mutation
Suppresses effect of an earlier mutation at a different site
Intragenic Suppressor Mutation
Suppresses effect of a mutation within the same gene
Intergenic Suppressor Mutation
Suppresses effect of a mutation in a different gene