lectures 3 and 4 - the human genome project
1/56
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
57 Terms
Range of sizes of human chromosomes
55×10^6bp to 250×10^6bp
smallest chromosome
Y
what other chromosome is X chromosome approx the same size as?
chromosome 8
what were the 4 main goals of the human genome project?
determine the sequence of the 3 billion base pairs
identify all human genes and their position
attempt to predict all genes’ function
use this information to understand disease, develop better medicine and see how humans compare to other species etc
What grant was provided to Francis Collins and team to carry out the HGP?
$200 million/year
Craig Venter
founder of Celera, the private competitor of the publicly funded HGP
how was work divided within the human genome project?
individual labs concentrated on a single chromosome
Why were robotic production lines required for the HGP?
to carry out the large number of PCR tests and sequencing reactions
sharing of information - HGP
The project was publicly funded so information was shared worldwide as it was found out on the internet for anyone to access - including Celera
HGP phase I
produce high res chromosomal maps by positioning genetic markers and creating libraries of BAC clones for sequencing
HGP phase II
sequence each BAC DNA
HGP phase III
assemble all sequences to produce final draft and annotate to identify genes
what is shotgun cloning?
breaking up DNA sequences into lots of small pieces then sequencing each and reassembling the sequence by using computers to look for regions of overlap
integrated maps of human chromosomes
genetic and physical maps were combined to put all BACs in the right order - logged as library, plate, row and column
What had to happen for the final phases of the HGP to occur?
Sequencing technology needed to improve. Other species’ genomes were sequenced in the wait for technology to become advanced enough
Old Sanger sequencing
radioactive 35S labelled DNA fragments were ran on a gel with 4 separate dideoxy reactions taking place (one for each base) then exposed to xray film
New Sanger sequencing
similar to PCR but fluorescently labelled bases act as terminators
HGP phase II and the Sanger method
advances in Sanger sequencing technology were taken advantage of, speeding up the process and reducing cost
how were DNA sequences assembled in phase II?
Computationally into ‘contig’ (set of overlapping DNA segments brought together)
what’s needed for one-fold coverage (phase II)?
approx 3 million separate sequencing reactions producing 1,000 bases each
draft sequence coverage/depth
4 fold coverage
complete/finished sequence coverage/depth
9 fold coverage
disadvantages of the clone-by clone approach (used by publicly funded international project)
very slow and expensive
advantages of the clone-by clone approach (used by publicly funded international project)
very effective at getting over regions of highly repetitive DNA sequences and it’s possible to retrieve clones later
Disadvantages of Celera’s approach
had to rely on public sequence databases and mapping information to assemble the sequence generated and then fill in gaps left by the repeats
Venter/Collins simultaneous publication
Both sides were only just able to produce a readable genome at the big announcement as Celera didn’t have enough sequence of their own and the public sequence was poor quality
White House announcement June 2000
the majority (80%) of the human genome had been sequenced in a working draft. Completion of 99.99% of the genome occurred in July 2003
genes of unknown function
20 years ago a figure was published saying 4061 (23.6%) of our genes’ functions were unclassified. This figure has barely changed since due to changes in public interest and direction of funding
How many people were sequenced during the human genome project?
4 in the public project and 1 at Celera. As the sequences were aligned, variants emerged
people studied in the international HapMap project
4 populations first studied (270 people total from Nigeria, Japan, China and Utah). More recently up to 1301 from across the world
purpose of the international HapMap project
to find the most common SNP variants in the world’s population. Reported locations of 1 million+ human SNPs published in Nature 2005
Moore’s law
describes the electronics field where chips get logarithmically faster and cheaper as time goes on - sequencing technologies are advancing at an even greater rate
Why are big sequencing studies becoming more routine?
Human genome sequencing is becoming very cheap and much faster. What once took 10 years can now take 24 hours
According to the 2022 draft ‘T2T-CHM13’, how many genes do humans have in their DNA, and how many of these are protein coding?
63,494 genes. 19,969 are predicted to code for proteins
What do non-coding genes do?
code for tRNAs, rRNAs, miRNAs and long non-coding RNAs (lncRNAs) involved in transcription
alternative mRNA splicing
humans have a relatively small number of genes. This process allows more protein isoforms to be produced per gene
What pathways are highly conserved across species?
pathways important to life eg ribosome proteins, tRNAs and ABC transporters (pump ions/hormones across membrane)
TBC1D3 function
regulates growth factors and has role in RAS-mediated cancers. Found only in humans
genes specific to humans
850 genes found with human specific features. 50+ entirely human specific genes found
What genes show evidence for fast/recent human evolution?
pathogen response, DNA & protein metabolism, reproduction, neuronal activity, skin pigmentation
How do most HIV strains enter cells?
using CCR5 as the main co-receptor (with CD4). Some people are naturally resistant as they’re homozygous for delta32 mutation in CCR5 gene
HIV has only been around since the 70s/80s - the prevalence of the CCR5 D32 variant would have required millenia - how?
the mutation may have been selected for in ancestral populations against earlier HIV-like epidemics or possibly smallpox
FOXP2
gene involved in human speech and language disorders
FOXP2 in humans and their ancestors
May have been key in human-specific language development - difference of 2 amino acids between chimps and humans/Neanderthals/Denisovans
human specific loss of olfactory (smell) genes
humans have far fewer functional olfactory genes than other species, suggesting that we rely more heavily on other senses eg sight and hearing for survival
linkage disequilibrium blocks
Collection of linked marker alleles on one chromosome over a short distance and tend to remain intact - not disrupted by meiotic recombination
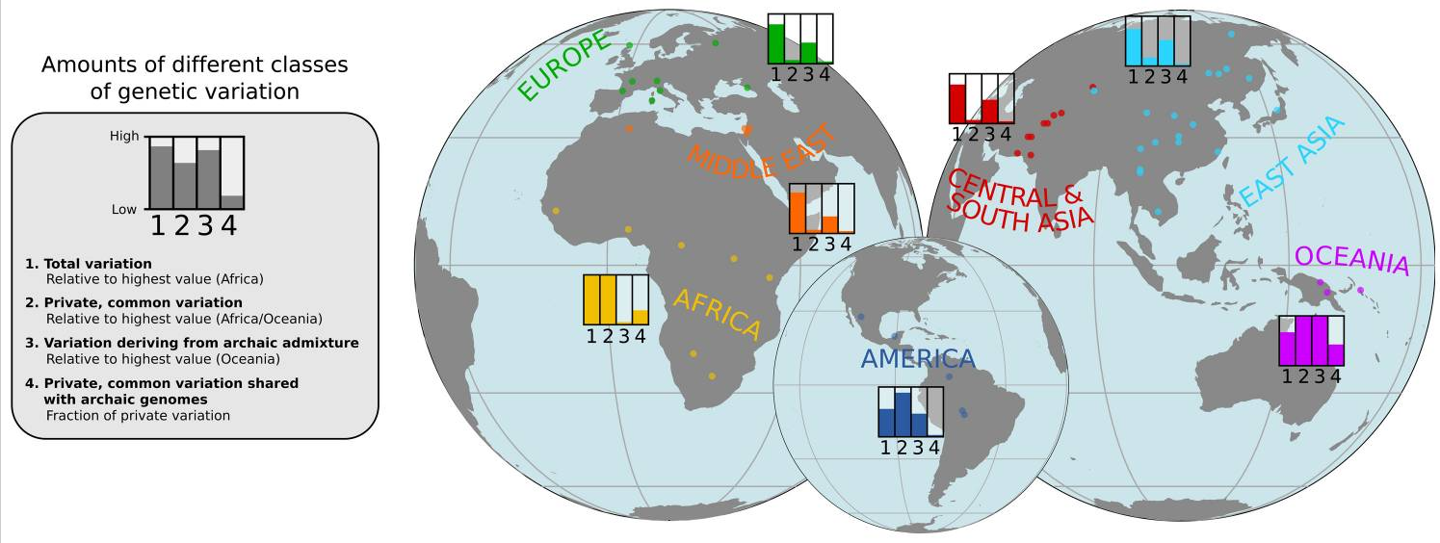
what is the genetic evidence for humans travelling out of Africa to populate the world?
African populations today are far more genetically diverse than the rest of the world as those that left were only a small population - example of a genetic bottleneck
humans as ancestors of Neanderthals and Denisovans
All populations have some form of genetic contribution from archaic ancestors. Approx 2 - 2.5% of a non-African genome derives from the genomes of Neanderthals and Denisovans
genetic differences affecting results of drugs (pharmacogenomics/genetics)
(in)activation by oxidative pathways
conjugation for excretion through kidney
target sensitivity
toxicity
disease mutation type
How could genome sequencing be used for personalised medicine?
to predict those who will respond well or badly (death/severe adverse reaction) and prescribe accordingly
screening pharmacogenomics and warfarin
screening for CYP2C9 and VKORC1 gene variants identifies those who should start on a low warfarin dose to reduce risk of internal bleeding
CYP2C19 variants and antidepressants
Antidepressant escitalopram affects brain serotonin levels and different gene variants affect the serum concentration and treatment success
(PegIFN-a)-2b/2a combined with RBV for Hep C treatment
Treatment really successful for individuals with CC SNP but not as much for TT SNP patients
most common mutation causing cystic fibrosis in Europeans
DF508 - no protein produced
Ivacaftor - CF drug
treats the 5% with the mutation that just damages protein function by helping to refold the protein therefore screening CF patients would find treatable cases. However would cost 200k/year/patient
Drug-Induced Liver Injury (DILI) and Flucloxacillin
SNP marker in complete LD with HLA-B*5701 immune cell surface protein vary significantly associated with DILI risk from antibiotic which affects 8.5/100,000. Screening would prevent prescription to those with the marker