Biology - Chapter 21: Recombinant DNA Technology
1/86
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
87 Terms
What does recombinant DNA technology involve?
The transfer of fragments of DNA from one organism, or species, to another
What problems are associated with introducing extracted chemicals into patients to treat a deficiency? (3)
Rejection
Risk of infection
Cost
Recombinant DNA
DNA that has sections of DNA from two species (by having the genes of one organism isolated, cloned and transferred to another)
Transgenic organism
An organism that carries recombinant DNA - it is the recipient of the transferred DNA
Alternative name for a transgenic organism
Genetically modified organism
Why can DNA be transferred between organisms and function normally?
The genetic code, as well as translation and transcription mechanisms, are universal
Outline the process of making a protein using the DNA technology of gene transfer and cloning (5)
Isolation of DNA fragments that have the gene for the desired protein
Insertion of the DNA fragments into a vector
Transformation - DNA fragments are transferred into suitable host cells
Identification using gene markers to identify the host cells that have successfully taken up the gene
Growth/cloning of the population of host cells
What feature of the structure of different proteins enable them to be separated by gel electrophoresis? (3)
Number of amino acids
Charge
Different R groups
3 methods of producing DNA fragments
Conversion of mRNA to cDNA using reverse transcriptase
Cutting fragments containing the desired gene from DNA using restriction endonucleases
Creating the gene in a gene machine (usually based on a known protein structure)
How is reverse transcriptase used to produce DNA fragments? (3)
mRNA of the desired protein is extracted from the cell that produces the protein
Reverse transcriptase is used to produce cDNA from the RNA
DNA polymerase forms the complementary strand of DNA by catalysing the condensation reaction between the aligned DNA nucleotides
cDNA
Complimentary DNA
How are restriction endonucleases used to produce DNA fragments?
It cuts the DNA double strand at the recognition sequence, leaving either blunt ends or sticky ends
Restriction endonucleases
Enzymes that cut up DNA
Why are there many types of restriction endonucleases?
Each one cuts a DNA double strand at a specific sequence of bases - the recognition sequence
Recognition sequence
A specific sequence of bases where restriction endonucleases cut the DNA strand
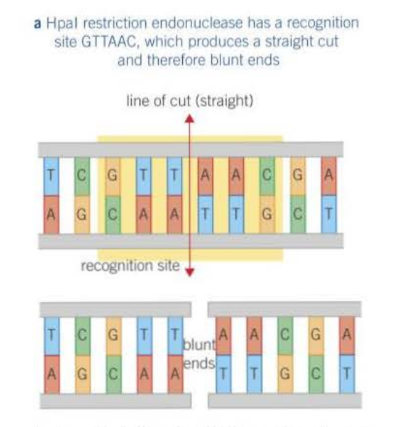
How do restriction endonucleases cut the DNA to form blunt ends?
The cut occurs between two opposite base pairs, leaving two straight edges.
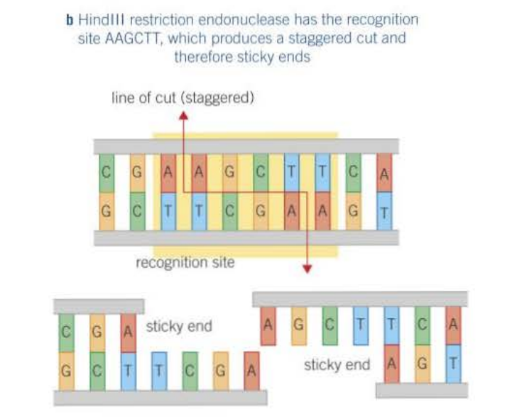
How do restriction endonucleases cut the DNA to form sticky ends?
The cut occurs in a staggered fashion, where each strand in the DNA has exposed, unpaired bases - the base sequences of the ends are opposites of each other i.e. palindromes
How is the gene machine used to produce DNA fragments? (9)
Desired seqquence of nucleotide bases is determined from the desired protein to be produced
mRNA codons are looked up, complementary DNA triplets are worked out
Desired nucleotide bases are fed into a computer
Sequence is checked for biosafety and biosecurity to ensure it meets international standards + ethical requirements
Computer designs oligonucleotides, which are assembled into the gene
Oligonucleotides are assembled into the required sequence by adding the nucleotide one at a time, producing a gene
The gene is replicated and the complementary DNA strand produced using the PCR
The gene is then inserted into a bacterial plasmid using its sticky ends
Genes are checked using standard sequencing techniques and those with errors are rejected
Oligonucleotides
Small, overlapping single strands of nucleotides
2 advantages of using the gene machine to produce DNA fragments
Any sequence of nucleotides can be produced in a very short time with great accuracy
The genes are free of introns and other non-coding DNA
In vivo
Transferring the fragments to a host cell using a vector
In vitro
Using the polymerase chain reaction
What happens if the same restriction endonuclease is used to cut DNA?
All the fragments produced will have ends complementary to one another, so the single-stranded end of any one fragment can be joined to the single-stranded end of any other fragment
How are the complementary bases of two sticky ends joined together?
DNA ligase is used to bind the phosphate-sugar backbone of the two sections of DNA and unite them as one
Why are sticky ends useful?
As long as the same restriction endonuclease is used, we can combine the DNA of one organism with that of any other organism
What must happen for transcription to take place?
RNA polymerase must attach to the DNA near a gene
Promoter
A region of DNA that acts as the binding site for RNA polymerase as well as transcription factors
Terminator
A region of DNA that releases RNA polymerase and ends transcription
What must be attached to a DNA fragment so that it will transcribe mRNA and produce a protein?
A promoter and terminator
Vector
Carries DNA into the other organism
Give an example of a vector
A plasmid
What restriction endonuclease is used to cut the plasmid?
The same one as the one used to cut the DNA fragment so the sticky ends will be complementary to each other
How are DNA fragments inserted into a vector? (2)
Restriction endonuclease used to cut plasmids to produce sticky ends - same one as the one used to cut DNA fragment
DNA ligase joins sticky ends of the DNA and plasmid together, forming recombinant DNA
How is DNA introduced into host cells? Why? (2+1)
Plasmids and bacterial cells are mixed with calcium ions
Temperature is also increased
This makes the bacterial membrane permeable so the plasmids can pass through into the cytoplasm
Transformation
The introduction of recombinant DNA into host cells
3 reasons why not all bacterial cells may possess the DNA fragments for the desired gene
Only a few bacterial cells will take up the plasmids when mixed together
Some plasmids close up without incorporating the DNA fragment
Some DNA fragments join together to form its own plasmid
How can you find out which bacterial cells have taken up plasmids? What is an issue associated with this? (3+1)
All bacterial cells are grown on a medium containing an antibiotic
Bacterial cells with plasmids have the gene for antibiotic resistance, so can break the antibiotic down and survive
The bacterial cells without the plasmids die
This includes bacterial cells that have taken up the plasmid and gene as well as cells that have taken up just the plasmid
3 examples of marker genes
Genes for antibiotic resistance
Genes that produce a fluorescent protein
Genes that produce an enzyme with an identifiable action
How is replica-plating used to identify cells with the recombinant DNA? (4)
In cells with the recombinant DNA, the gene for resistance to another antibiotic is cut due to the inserted DNA
Replica-plating is used to replicate the arrangement of the bacteria colonies
One of the plates is grown with a sample with that antibiotic
The colonies that die have recombinant DNA, so are isolated and used
How are fluorescent markers used to identify cells with the recombinant DNA? (3)
Gene that codes for a fluorescent protein is transferred to the plasmid
Gene to be cloned is inserted in the middle of the gene for the fluorescent protein, so cells with the recombinant DNA do NOT glow
The cells that do glow do NOT have the recombinant DNA, so are discounted
How is enzyme-markers used to identify cells with the recombinant DNA? (3)
Gene that codes for an enzyme is transferred to the plasmid
Gene to be cloned is inserted in the middle of the gene for the enzyme, so cells with the recombinant DNA do NOT change colour as a result of the enzyme’s action
The cells that do change colour do NOT have the recombinant DNA, so are discounted
Polymerase Chain Reaction
A method of copying fragments of DNA
Why is PCR rapid and efficient?
It is automated
3 things that need to be added to the DNA fragment in PCR
DNA polymerase
Primers
Nucleotides
Describe the process of PCR (4)
Mixture of DNA polymerase, DNA nucleotides and primers
Temperature is heated to 95°C to break the hydrogen bonds between the DNA strands, causing the strands to separate
Temperature is cooled to 55°C so the primers bind to the ends of the DNA strands (and nucleotides attach by complementary base pairing)
Temperature is heated to 72°C so DNA polymerase joins the adjacent nucleotides together by catalysing the condensation reaction between them
What happens to the number of DNA molecules after a cycle of DNA polymerase?
It doubles
2 advantages of in vitro cloning
It is extremely rapid
It does not require living cells - only need base sequence of DNA
4 advantages of in vivo cloning
Useful if want to introduce a gene into another organism
Involves almost no risk of contamination
It is very precise and only cuts out specific genes
Produces transformed bacteria that can be used to produce large amounts of gene products
2 ways to find out where a gene is located
By using labelled DNA probes or DNA hybridisation
2 most commonly used probes
Radioactively-labelled probes: use isotope 12P - probe identified with X-ray film exposed by radioactivity
Fluorescently-labelled probes: emit light under certain conditions
When does DNA hybridisation take place?
When a section of DNA/RNA is combined with a single stranded section of DNA which has complementary bases
What might DNA probes be used to screen for? (3)
Heritable conditions, drug responses or health risks
What are the benefits of genetic screening? (3)
People at a greater risk of developing a disease can make informed decisions about their lifestyle and future treatment
They can be checked more regularly, so may be diagnosed earlier and have a better chance of successful treatment
Personalised medicine: people can receive medical advice and treatment specific to their genotype
Genetic counselling
A form of social work where people are provided with advice and information to enable them to make personal decisions about themselves or their offspring
3 things genetic screening can detect with regards to cancer
Oncogene mutations: can determine type of cancer person has and so most effective drug / radiotherapy available
Gene changes that predict which patients are more likely to benefit from certain treatments and have the best chance of survival
A single cancer cell among normal cells, so can tell whether someone is at risk of relapsing
Why might you use the gene machine over using reverse transcriptase / restriction endonuclease?
It is faster to use the gene machine than all the enzyme-catalysed reactions
Why would you not use restriction endonucleases to obtain DNA fragments when the fragments are going to be inserted into bacterial plasmids? (2)
Human DNA contains introns
The bacteria cannot splice pre-mRNA
Why is it important that inserted genes are only expressed in specific places? (2)
So protein can be harvested - it may be easy to extract
Producing the protein in other cells / elsewhere, it may damage other cells, causing harm
When trying to carry out PCR for a sample of genetic material, why might you need to produce a variety of primers?
The DNA/RNA base sequences differ, so different complementary primers are needed
Why might bacteria not be able to produce every human protein? (3)
Do not have required transcriptional factors, so cannot carry out transcription and produce mRNA
Cannot splice pre-mRNA, so cannot remove introns
Do not have Golgi apparatus, so cannot modify proteins
Variable number tandem repeats
DNA bases which are non-coding
VNTRs
Variable Number Tandem Repeats
What is the probability of two individuals having identical VNTR sequences? Why?
Very low - for each individual, the number and length of VNTRs has a unique pattern. This is different for all individuals except identical twins.
How does relation affect the pattern of VNTR sequences?
The more closely-related two individuals are, the more similar the VNTRs will be
What is gel electrphoresis used for?
To separate DNA fragments according to their size
In gel electrophoresis, how does the size of the fragment affect the speed they move at? Why?
The larger the fragment, the slower they move - this is due to the resistance of the agar gel
What must be done before larger genes and whole genomes go through gel electrophoresis?
They must be cut into smaller fragments by restriction endonucleases
What is genetic fingerprinting used for? (4)
Determining genetic relationships and the genetic variability in a population
Forensic science
Medical diagnosis
Plant and animal breeding
Why is genetic fingerprinting used to determine genetic relationships and the genetic variability in a population? (3)
The more closely related individuals are, the closer the resemblance of their genetic fingerprints
Relatives will share some bands of their genetic fingerprints
A population that has members with very similar genetic fingerprints have little genetic diversity
Why is genetic fingerprinting used in forensic science?
DNA found at the crime scene can be analysed and compared to identify someone at the scene of the crime
Why is genetic fingerprinting used in medical diagnosis? (2)
Can be used to determine if someone has base sequence / alleles responsible for a genetic disorder, the probability of them developing and when
Can be used to identify the nature of a microbial infection by comparing fingerprint of the microbe with other similar pathogens
Why is genetic fingerprinting used in plant and animal breeding? (2)
Can be used to select individuals for breeding if they have a desirable allele / prevent individuals with undesirable alleles from breeding
Can be used to determine the paternity of an animal, establishing its pedigree
Describe how genetic testing / genetic fingerprinting / DNA screening is carried out. (7)
DNA extracted from sample
PCR used to amplify DNA sample
DNA cut into segments using restriction endonucleases
DNA fragments separated using electrophoresis according to size / length
DNA strands separated by an alkali to form single strands
Labelled DNA probes are applied and anneal to the sample (by DNA hybridisation)
Mutations identified using X-ray film due to radioactivity OR bands compared to DNA sample (e.g. one with mutations)
DNA primers
Short lengths of single-stranded DNA
Why are DNA primers added in PCR? (3)
To mark the beginning and/or ends of the part of DNA needed
So enzymes / nucleotides can attach
To keep the DNA strands apart
What is special about the DNA polymerase used in PCR?
It is thermostable
Why is it important that the DNA polymerase enzyme used in PCR is thermostable?
It would not be denatured - in PCR, it must be heated to 95°C, so it must be able to withstand high temperatures
Why is PCR needed before carrying out genetic fingerprinting?
Only small sample of DNA is obtained / would need more DNA for analysis - PCR gives many copies
What must be done if a DNA sample is contaminated before obtaining a genetic fingerprint? Why?
Need to identify “required” DNA from rest as other DNA is present
What is an advantage of including promoter DNA when inserting recombinant DNA for a desired gene into an organism?
It ensures that the desired gene is only expressed in the needed organ and not elsewhere
How are DNA probes used to identify what cells contain a gene? (6)
Extract DNA
Add restriction endonucleases
Separate fragments using gel electrophoresis
Treat DNA using an alkali to separate the strands and form single strands
Probe anneals to gene
Use X-ray film to show bound probe
DNA probe
A short single strand of DNA with bases complementary to the base sequence of DNA
How do DNA probes work? (3 - brief answer)
Probes are single stranded
They have a specific base sequence complementary to the DNA
They bind to the DNA and can be identified
Why would it be useful to identify the nature of a disease using genetic fingerprinting? (3)
To see if pathogen is resistant to treatment so can prescribe an effective one (e.g. antibiotics)
To see if a vaccine would work and what vaccine to use so can reduce the spread
To test others and see if they have the same strain so the spread of the disease can be controlled + people can be vaccinated/treated
When inserting a plasmid with recombinant DNA into an organism, why should a marker gene be used?
Because not all cells will successfully take up the plasmid so can identify the ones that did
What are vectors used for in genetic engineering?
To carry genes from one organism to another
What features of DNA sequences enable them to be separated by gel electrophoresis?
Number of nucleotides
(Negative) charge