tutorial 2: Western Blot Day 1 protein extraction students (copy)
1/27
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
Western Blot
detection of proteins
Southern Blot:
detection of DNA (Named after its inventor, Edwin Southern)
Northern Blot
detection of RNA
Eastern Blot:
using different types of probes to detect post-translational modifications of proteins
what are the steps of experiment 1
Cellular proteins will be isolated from two different E. coli strains, EMG 26 K-12 lac- (i+z+y-) and EMG 9 K-12 lac- (i-z+y+). One of these strains produces the beta-galactosidase enzyme.
This protein extract will be subject to SDS-PAGE to produce a protein profile based on molecular weight.
Part of the gel will be stained with Coomassie blue to show the presence of all the proteins.
The proteins on the remainder of the gel will be transferred to nitrocellulose filter by electrophoretic transfer.
Visualization of the beta-galactosidase protein on the nitrocellulose filter will be carried out by absorption with an anti-beta-galactosidase monoclonal antibody followed by detection with an anti-IgG alkaline phosphatase conjugate.
describe the lac operon
The lac operon contains three enzyme-coding structural genes (lacZ, lacY, lacA) and three regulatory elements (repressor I, promoter, operator). The enzymes work together to allow E. coli to digest the disaccharide lactose, and the regulatory elements control the transcription of these enzymes
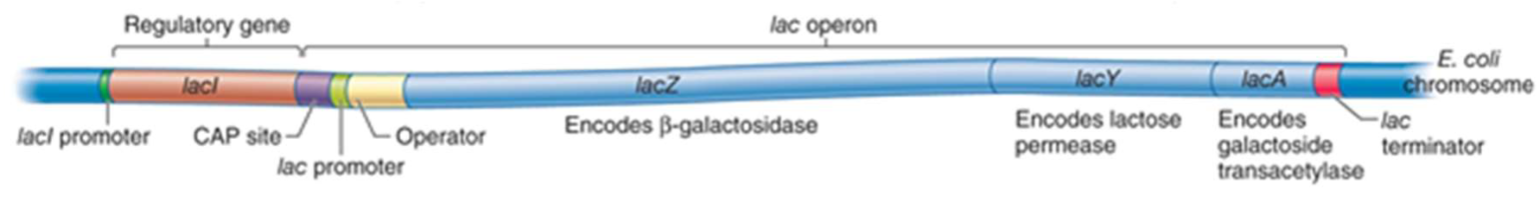
lacZ
encodes beta-galactosidase, an enzyme that takes lactose as a substrate and cleaves it into the monosaccharides galactose and glucose. This is the first reaction necessary for the breakdown of lactose
lacY
encodes lactose permease, a membrane-bound transport protein that pumps lactose into the cell. In the cell, lactose is hydrolyzed by β-galactosidase (or lactase) to glucose and galactose
lacI
gene codes for the repressor, a protein which binds to the operator and inhibits transcription
lacA
codes for Beta-galactoside transacetylase (lacA), an enzyme that adds acetyl groups to lactose and other galactose-containing sugars. The role of this enzyme in lactose digestion is not well defined
operon
structural genes, together with promotor and operator or activator-binding site
CAP
CAP – catabolite activator protein is a transcription activator (increases the rate of transcription)
promoter
a region of DNA that initiates transcription of a particular gene. An RNA polymerase must bind to the promoter region to begin mRNA transcription
operator and repressor protein
contains a binding site for the repressor protein I. When the repressor protein is bound to the operator, RNA polymerase cannot bind to the promoter.
the operator region binds repressor proteins (e.g. lac repressor; lacI, upstream of lac operon).
an operator is a segment of DNA to which a transcription factor binds to regulate gene expression
A repressor protein is a DNA or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator.
what happens when lactose is present vs when it is not present?
Enzymes normally not produced unless lactose present; lac repressor inhibits transcription in the absence of lactose
– Recall that lacZ encodes beta galactosidase, which converts lactose to allolactose, the inducer of the operon.
– Allolactose interacts with the lac repressor and causes the repressor to change to an inactive shape that is unable to bind to an operator site.
– The inactivated repressor leaves the DNA and transcription occurs
function of Tris-HCl buffer, pH=6.8 in experiment
a buffering agent used for pH regulation
function of lysis buffer, pH=6.8 in experiment
Lysis buffers differ in their ability to solubilize proteins, with those containing sodium dodecyl sulfate (SDS) and other ionic detergents considered to be the harshest and therefore most likely to give the highest yield.
components of the lysis buffer
lysis buffer also contains glycerol, EDTA and 2% mercaptoethanol
function of glycerol
makes the sample more dense than the sample buffer, so the sample will remain in the bottom of a well rather than float out”
function of EDTA
chelating agent and its role in the lysis buffer is to reduce oxidation damage and to chelate metal ions (and this inhibits some proteases)
function of mercaptoethanol
is a reducing agent that reduces (disrupts) disulphide bridges in proteins, which is necessary for separation by size.
Disulphide bonds can form between thiol groups of cysteine residues.
Breaking S-S bonds may lead to disruption of tertiary and quaternary structure of proteins, resulting in protein monomers
function of running dye
“The dye allows the investigator to track the progress of the electrophoresis”.
Bromophenol blue has a slight negative charge and will migrate the same direction as DNA (or other negatively charged molecules), allowing the user to monitor the progress of molecules moving through the gel.
The rate of migration varies with gel composition.”
function of SDS (Sodium dodecyl sulfate)
anionic detergent
cell lysis
the purpose of the SDS detergent is to take the protein from its native shape, and open it up into a linear piece and coat them with a uniform negative charge - proteins separated only on basis of size and not charge
This will allow it to run more efficiently down a gel and it will be easier to compare the linear pieces of proteins rather than the native shapes of protein
describe SDS
how does it bind
how does it interact with proteins
The SDS has a hydrophobic tail that interacts strongly with protein (polypeptide) chains
The hydrophobic portion of SDS interacts with hydrophobic regions of proteins.
SDS (negatively charged) binds to the protein structure in the positively charged surface areas and then alters the native binding to the neighboring hydrophobic residues
SDS binds fairly uniformly to the linear proteins (around 1.4g SDS/ 1g protein), meaning that the charge of the protein is now approximately proportional to its molecular weight.
SDS also disrupts the forces that contribute to protein folding (tertiary structure), ensuring that the protein is not only uniformly negatively charged, but linear as well.
what is the number of SDS molecules that bind to a protein proportional to?
the number of amino acids that make up the protein
Detergents
class of molecules whose unique properties enable manipulation (disruption of conformation) of hydrophobic–hydrophilic interactions among molecules in biological samples.
In biological research, detergents are used to lyse cells (release soluble proteins), solubilize membrane proteins and lipids
Denaturing detergents can be anionic such as sodium dodecyl sulfate (SDS) or cationic.
These detergents totally disrupt membranes and denature proteins by breaking protein– protein interactions
what does SDS do? what determines migration?
When SDS is used with proteins, all of the proteins become negatively charged by their attachment to the SDS anions
SDS confers a negative charge to the polypeptide in proportion to its length.
Denatured polypeptides become rods of negative charge with equal charge densities per unit length
Therefore, migration is determined by molecular weight, rather than by the intrinsic charge of the polypeptide.
what protein structures does SDS affect?
SDS breaks up the two- and three-dimensional structure of the proteins by adding negative charge to the amino acids.
Since like charges repel, the proteins are more-or-less straightened out, immediately rendering them functionless.
Some quaternary structure may remain due to disulfide bonding (covalent) and due to covalent and noncovalent linkages to other types moleculeseecules