BIOL121: Exam 3
1/179
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
180 Terms
() are viruses that infect bacteria
(bacteriophages) are viruses that infect bacteria
Prokaryotic viruses must bind to a () and cross a () or () for gram - hosts to not damage host cell
They exit through () and usually () host cells
Prokaryotic viruses must bind to a (host cell receptor) and cross a (cell wall) or (2 membranes) for gram - hosts to not damage host cell
They exit through (cell wall) and usually (lyse/bursts) host cells
Before it enters either of its 2 life cycles, bacteriophages attach to () using () and () through cell wall into cytoplasm like shooting the bacteria
Before it enters either of its 2 life cycles, bacteriophages attach to (host cell proteins) using (cell surface receptors) and (injects genome) through cell wall into cytoplasm like shooting the bacteria
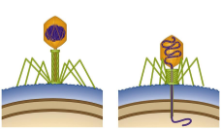
In the () cycle, the bacteriophage quickly replicates, killing host cell'
It can be broken down into 3 further steps after entering the cell: use cell components to synthesize (), assemble (), and exit through () or ()
Lysis makes protein to break () and bursts host cell to release progeny phage while slow release has () slip individual progeny out through cell envelope
In the (lytic) cycle, the bacteriophage quickly replicates, killing host cell
It can be broken down into 3 further steps after entering the cell: use cell components to synthesize (capsids), assemble (progeny phages), and exit through (lysis) or (slow release)
Lysis makes protein to break (peptidoglycan) and bursts host cell to release progeny phage while slow release has (filamentous phages) slip individual progeny out through cell envelope
() are viruses that infect E.coli and have 2 forms either () or () which both have linear dsDNA
The difference is () is virulent/lytic/intemperate while () is lysogenic/temperate
(Coliphages) are viruses that infect E.coli and have 2 forms either (T4) or (lambda) which both have linear dsDNA
The difference is (T4) is virulent/lytic/intemperate while (lambda) is lysogenic/temperate
Bacteriophage T4 has a complicated structure with () genes, () different capsid proteins, () outer membrane protein receptor, and a long tail for ()
Bacteriophage T4 has a complicated structure with (170) genes, (10) different capsid proteins, (OmpC) outer membrane protein receptor, and a long tail for (injecting DNA)
The T4 phage life cycles takes approximately 25 minutes and consists of:
() and ()
Early (): () synthesis and arrest of ()
()
Late () and formation of ()
() and releases newly formed viruses
The T4 phage life cycles takes approximately 25 minutes and consists of:
(Adsorption) and (penetration)
Early (mRNA synthesis): (enzyme) synthesis and arrest of (host gene)
(DNA replication)
Late (mRNA synthesis) and formation of (new viral particles)
(Host cell bursts open) and releases newly formed viruses
Upon entry, phage T4 genome forms a () and early genes transcribed that take control of () and destroy ()
It uses cell () to replicate genome in () that makes continuous replication of many copies of genome
Upon entry, phage T4 genome forms a (circle) and early genes transcribed that take control of (cell) and destroy (cell chromosome)
It uses cell (nucleotides) to replicate genome in (“rolling circle replication”) that makes continuous replication of many copies of genome

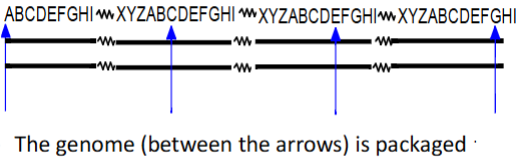
T4 progeny genomes are linked in () which is () linked together and cut with an () so individual linear genomes have slight ()
T4 progeny genomes are linked in (concatemer) which is (several genomes) linked together and cut with an (offset) so individual linear genomes have slight (overlaps)
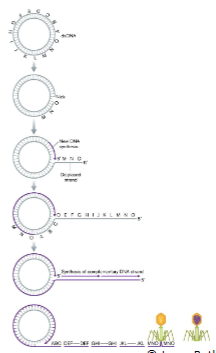
Each packaged DNA in the virus head is () than the complete T4 phage chromosome and sequence begins and ends at () points in different virions
Each packaged DNA in the virus head is (slightly longer at 103%) than the complete T4 phage chromosome and sequence begins and ends at (different) points in different virions
T4 phage particles () as late genes are transcribed the () polymerizes around progeny DNA, () and () made, and () assemble together
() protein made to destroy cell wall and release progeny
T4 phage particles (self-assemble) as late genes are transcribed the (head) polymerizes around progeny DNA, (tail fibers) and (long tail) made, and (head, tail, and tail fibers) assemble together
(Lysis) protein made to destroy cell wall and release progeny
() phages are phages that maintain a () relationship with the host cell where they stay with the host
These types of phages are capable of () where they integrate viral genome () into host DNA and become a ()
(Temperate) phages are phages that maintain a (stable) relationship with the host cell where they stay with the host
These types of phages are capable of (lysogeny) where they integrate viral genome (prophage) into host DNA and become a (lysogen)
Prophage DNA is mostly () and doesn’t enter () until induced by damaged DNA or a bad environment
Only () lysogenic virus of a particular type can be present in host cell
Prophage DNA is mostly (dormant) and doesn’t enter (lytic cycle) until induced by damaged DNA or a bad environment
Only (a single) lysogenic virus of a particular type can be present in host cell
Coliphage lambda encodes () genes, has a linear dsDNA with (), and has a 12 nucleotide complementary ss regions
The lambda phage receptor in E. coli is ()
Coliphage lambda encodes (50) genes, has a linear dsDNA with (cohesive ends/cos sites), and has 12 nucleotide complementary ss regions
The lambda phage receptor in E. coli is (porins)
Lambda phage DNA in phage head is () but () in host cell
() combine in host to give circular configuration at ligated cos site
cos recognition sites determine where the genome is () and ()
Lambda phage DNA in phage head is (ds and linear) but (circular) in host cell
(Tandem repeating ends) combine in host to give circular configuration at ligated cos site
cos recognition sites determine where the genome is (cut) and (packaged)
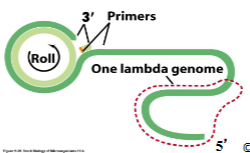
Lambda phage replication begins with () at the beginning it initiates at () site in () replication
() is when there’s continuous replication of many copies of genome and lambda has long chains of () genomes where cutting occurs at (), generating the linear form for packaging
Lambda phage replication begins with (theta) at the beginning it initiates at (ori) site in (bidirectional) replication
(Rolling circle replication) is when there’s continuous replication of many copies of genome and lambda has long chains of (concatenated) genomes where cutting occurs at (cos sites), generating the linear form for packaging
() is when viruses can integrate their viral genome into bacterial chromosome at () sites
When entering () cycle, bacterial genes () to viral attachment () sites are sometimes picked
(Lysogeny) is when viruses can integrate their viral genome into bacterial chromosome at (att) sites
When entering (lytic) cycle, bacterial genes (adjacent) to viral attachment (att) sites are sometimes picked
T/F: All infections are fatal
F; not all infections are fatal
Culturing bacteriophage
Dilute phage mixed with () on top and then ()
Make serial dilutions
Pour mixture onto petri dishes containing bottom agar
Plates incubated overnight
() grows
() infects () cells and multiply
() containing virions formed
Culturing bacteriophage
Dilute phage mixed with (agar) on top and then (E. coli)
Make serial dilutions
Pour mixture onto petri dishes containing bottom agar
Plates incubated overnight
(E. coli) grows
(Phage) infects (E. coli) cells and multiply
(plaques) containing virions formed
On a plate the () area is growth of E. coli as a lawn while () areas are () E. coli killed by phage
() are formed upon lysis of E. coli cell and is measured as ()
On a plate the (cloudy) area is growth of E. coli as a lawn while (cleared) areas are (plaques) E. coli killed by phage
(Plaques) are formed upon lysis of E. coli cell and is measured as (plaque forming units/pfu)
Bacteria - Host Interactions
() is living together
() is one benefits while the other isn’t affected
() is when both organisms benefit
() is when one benefits and the other is harmed
Bacteria - Host Interactions
(Symbiosis) is living together
(Commensalism) is one benefits while the other isn’t affected
(Mutualism) is when both organisms benefit
(Parasitism) is when one benefits and the other is harmed
T/F: Normal microbiota and hosts are usually parasitic
F; Normal microbiota and hosts are usually mutually beneficial
Normal microbiota often prevents () by pathogens and bacterial products are () to host
Normal microbiota often prevents (colonization) by pathogens and bacterial products are (beneficial) to host
Steps of bacterial infections:
() and () host tissues
() host defenses
Acquire () from host
() in host
() to new host
Steps of bacterial infections:
(Attach to) and (invade) host tissues
(Suppress) host defenses
Acquire (nutrients) from host
(Reproduce) in host
(Transmit) to new host
() is when a pathogen grows and multiplies within or on another organism and in most cases it is removed by immue system
() is due to the presence and multiplication of pathogens when part or all of the host can’t perform normal functions
(Infection) is when a pathogen grows and multiplies within or on another organism and in most cases it is removed by immue system
(Infectious disease) is due to the presence and multiplication of pathogens when part or all of the host can’t perform normal functions
() is an agent of disease while () is the process by which microbes cause disease in host
() like bacteria, fungi, viruses, and protozoa colonize and harm the host
(Pathogen) is an agent of disease while (pathogenesis) is the process by which microbes cause disease in host
(Parasites) like bacteria, fungi, viruses, and protozoa colonize and harm the host
() have the ability to penetrate host defenses and their whole life exists to be pathogens while () can only cause disease in compromised hosts
(Primary pathogens) have the ability to penetrate host defenses and their whole life exists to be pathogens while (opportunistic pathogens) can only cause disease in compromised hosts
Some microbes enter a () during infection and can remain dormant for years
Some microbes enter a (latent state) during infection and can remain dormant for years
() is the ability of an organism to cause disease and is defined in terms of () which is how easily an organism causes disease and () which is how severe the disease is
(Pathogenicity) is the ability of an organism to cause disease and is defined in terms of (infectivity) which is how easily an organism causes disease and (virulence) which is how severe the disease is
Virulence of organism can be measued in 2 ways: () which is the number of organisms to colonize 50% of hosts and () which is the number of organisms to kill 50% of hosts
For both cases () is worse
Virulence of organism can be measued in 2 ways: (ID50) which is the number of organisms to colonize 50% of hosts and (LD50) which is the number of organisms to kill 50% of hosts
For both cases (lower) is worse
The route an organism takes to spread disease is known as the ()
The route an organism takes to spread disease is known as the (infection cycle)
() are individual characteristics of pathogens that allow pathogens to invade hosts and cause disease
All () () the disease producing ability of pathogens, they don’t make the pathogen more lethal, they just help the pathogen cause disease
(Virulence factors) are individual characteristics of pathogens that allow pathogens to invade hosts and cause disease
All (virulence factors) (enhance) the disease producing ability of pathogens, they don’t make the pathogen more lethal, they just help the pathogen cause disease
() encode factors allowing pathogen to invade host
() are sections of genome that contain multiple virulence factors, often encoding related functions such as protein secretion systems and toxin production
(Virulence genes) encode factors allowing pathogen to invade host
(Pathogenicity islands) are sections of genome that contain multiple virulence factors, often encoding related functions such as protein secretion systems and toxin production
() can be transferred as a block from other organisms through (), are often flanked by phage or plasmid genes, and have () content different from the rest of the genome
(Pathogenicity islands) can be transferred as a block from other organisms through (horizontal gene transfer), are often flanked by phage or plasmid genes, and have (GC) content different from the rest of the genome
Bacteria must adhere to () if they want to colonize using either () which are hollow fibrils with tips to bind host cells or () which are surface proteins that bind host cells/microbial factor that helps attachment
Bacteria must adhere to (host tissue) if they want to colonize using either (pili/fimbriae) which are hollow fibrils with tips to bind host cells or (adhesins) which are surface proteins that bind host cells/microbial factor that helps attachment
() pili adhere to mannose residues on host cell surfaces and are considered static
(Type I) pili adhere to mannose residues on host cell surfaces and are considered static
() pili assemble on cell surface and are considered dynamic, continuously assembling and disassembling
(Type IV) pili assemble on cell surface and are considered dynamic, continuously assembling and disassembling
() pili subunits are made in the cytoplasm, then secreted to the periplasm
() chaperones the subunits to ()
() forms the pore for the pili units to assemble
All the other proteins assemble in order: PapG-PapF-PapE-PapA, then the cap PapH is added
(Type I) pili subunits are made in the cytoplasm, then secreted to the periplasm
(PapD) chaperones the subunits to (PapC)
(PapC) forms the pore for the pili units to assemble
All the other proteins assemble in order: PapG-PapF-PapE-PapA, then the cap PapH is added
() pili is involved in gliding or twitching motility often nicknamed “spider man motility”
(Type IV) pili is involved in gliding or twitching motility often nicknamed “spider man motility”
Bacteria can use () that mediate binding to host tissues like:
Streptococcus pyogenes which causes strep throat ()
Bordetella pertussis which causes whooping cough ()
Bacteria can use (non-pilus adhesions) that mediate binding to host tissues like:
Streptococcus pyogenes which causes strep throat (M protein)
Bordetella pertussis which causes whooping cough (pertactin)
Bacteria can attach to surfaces, forming a () which are often resistant to ()
Bacteria can attach to surfaces, forming a (biofilm) which are often resistant to (antibiotics)
Bacteria produce () to help them invade the host, damage host cells, or evade the immune system
Many microbes secrete () after attachment which kills host cells and releases nutrients
Bacteria produce (toxins) to help them invade the host, damage host cells, or evade the immune system
Many microbes secrete (exotoxins) after attachment which kills host cells and releases nutrients
() are non-protein toxic compounds that are hyper-active and harm immune systems
They are present in () bacteria as part of the () released then the cell breaks down
(Endotoxins) are non-protein toxic compounds that are hyper-active and harm immune systems
They are present in (gram negative) bacteria as part of the (lipopolysaccharide) released when the cell breaks down
There are 5 categories of protein ():
() disruption which causes host cell () leakage
Block () which targets eukaryotic ()
Block () pathways
() overactivate ()
() cleave ()
There are 5 categories of protein (exotoxins):
(cell membrane) disruption which causes host cell (membrane) leakage
Block (protein synthesis) which targets eukaryotic (ribosomes)
Block (2nd messenger) pathways
(Superantigens) over activate (immune system)
(Proteases) cleave (host proteins)
() is in the category 1 of toxins and forms a (), 7 member pre in target cell membranes disrupting () by forming () in the membrane
The () is produced by Staphylococcus aureus
(Alpha toxin) is in the category 1 of toxins and forms a (transmembrane), a 7 member pore in target cell membranes disrupting (red blood cells) by forming (pore) in the membrane
The (hemolytic alpha toxin) is produced by Staphylococcus aureus
() is in the category 2 of exotoxins and disrupts () by destroying 28S rRNA found in eukaryotic ()
The () is produced by Shigella flexneri and E. coli
(Shiga toxin) is in the category 2 of exotoxins and disrupts (protein synthesis) by destroying 28S rRNA found in eukaryotic (ribosomes)
The (shiga toxin) is produced by Shigella flexneri and E. coli
() is a 2 component toxin which is made of the () which has toxic activity and the () which binds to host cell and delivers () to ()
() is also known as the ADP ribosyltransferase and is part of Diphtheria and Cholera toxins
(AB toxin) is a 2 component toxin which is made of the (B subunit) which has toxic activity and the (A subunit) which binds to host cell and delivers (A) to (cytoplasm)
(A subunit) is also known as the ADP ribosyltransferase and is part of Diphtheria and Cholera toxins
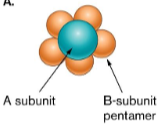
() is only made by () bacteria, is a component of outer membrane, and contains ()
() is released as the bacteria dies which can cause toxic shock and massively activates host () repsonse
(Lipopolysaccharide) is only made by (gram negative) bacteria, is a component of outer membrane, and contains (endotoxins)
(Lipid A) is released as the bacteria dies which can cause toxic shock and massively activates host (inflammatory) response
People need to be careful when administering treatments against infections because too much treatment can cause massive release of ()
People need to be careful when administering treatments against infections because too much treatment can cause massive release of (endotoxins)
Many pathogens uses specific protein secretion pathways to deliver ()
The proteins may not kill the cell but redirect () to benefit the microbe
Many pathogens uses specific protein secretion pathways to deliver (toxins)
The proteins may not kill the cell but redirect (host signaling pathways) to benefit the microbe
() is the general secretion pathway and consists of 3 portions (), () or membrane fusion protein, and () channel protein
() is secreted through this system
(Type I secretion system) is the general secretion pathway and consists of 3 portions (ATP binding cassette protein), (periplasmic protein) or membrane fusion protein, and (outer membrane) channel protein
(Hemolysin alpha toxin) is secreted through this system
() uses pilus like mechanisms to push proteins out of the bacterial cell similar to type IV pili and is modified to extend and retract for secreting proteins
(Type II secretion system) uses pilus like mechanisms to push proteins out of the bacterial cell similar to type IV pili and is modified to extend and retract for secreting proteins
() is like a molecular syringe to inject proteins from bacterial into host cytoplasm in one step
It is essential for () pathogens like E. coli, salmonella, yersinia pestis, and pseudomonads
(Type III secretion system) is like a molecular syringe to inject proteins from bacterial into host cytoplasm in one step
It is essential for (gram negative) pathogens like E. coli, salmonella, yersinia pestis, and pseudomonads
() uses proteins that resemble conjugation machinery to secrete proteins or DNA
It can extend and retract to move proteins into or out of the cell
(Type IV secretion system) uses proteins that resemble conjugation machinery to secrete proteins or DNA
It can extend and retract to move proteins into or out of the cell
() are injected proteins from the () which induces tight attachment to host, causes host to engulf bacteria, induce () in cell, suppresses host immunity, and causes diarrhea
(Type III effectors) are injected proteins from the (type III secretion system) which induces tight attachment to host, causes host to engulf bacteria, induce (actin rearrangement) in cell, suppresses host immunity, and causes diarrhea
() are compounds that are produced by one species of microbe that can inhibit the growth or kill other microbes
() found () in 1929 and it was again rediscovered by () in 1940
(Antibiotics) are compounds that are produced by one species of microbe that can inhibit the growth or kill other microbes
(Alexander Fleming) found (penicillin) in 1929 and it was again rediscovered by (Howard Florey) in 1940
() discovered () which is metabolized by the body into sulfanilamide
Sulfanilamide is an analog of para-aminobenzoic acid which is a () precursor
(Gerhard Domagk) discovered (sulfa drugs) which is metabolized by the body into sulfanilamide
Sulfanilamide is an analog of para-aminobenzoic acid which is a (folic acid) precursor
Sulfa drugs stop bacterial growth by inhibiting conversion of () into ()
Bacteria’s use () to make () and if the can’t make () they can’t grow
Sulfa drugs stop bacterial growth by inhibiting conversion of (PABA) into (folic acid)
Bacteria’s use (PABA) to make (folic acid) and if they can’t make (folic acid) they can’t grow
Antibiotics must affect () but not patients
Drugs should only affect () like () and many can have side effects at () or cause ()
Antibiotics must affect (infectious organism) but not patients
Drugs should only affect (microbial physiology) like (peptidoglycan) and many can have side effects at (high concentrations) or cause (allergic response)
() antimicrobials are effective against many species while () are effective against few or a single species
(Broad spectrum) antimicrobials are effective against many species while (narrow spectrum) are effective against few or a single species
() kill target organism while () prevents the growth of target organism while the immune system kills the infection
(Bactericidal) antibiotics kill target organism while (bacteriostatic) prevents the growth of target organism while the immune system kills the infection
() drugs only affect growing cells by inhibiting ()
It is only effective if organism is building a new () and patients must take the antibiotic until all cells leave () phase
(Bacteriostatic) drugs only affect growing cells by inhibiting (cell wall synthesis)
It is only effective if organism is building a new (cell wall) and patients must take the antibiotic until all cells leave (stationary) phase
() is the lowest concentration of an antibiotic that prevents growth and it varies for different bacterial species
No colonies formed is called ()
(Minimal inhibitory concentration) is the lowest concentration of an antibiotic that prevents growth and it varies for different bacterial species
(Minimal lethal concentration) is the lowest concentration of an antibiotic that results in the death of bacterial species
() disk diffusion test tests bacterial sensitivity to multiple antibiotics
(Kirby-Bauer) disk diffusion test tests bacterial sensitivity to multiple antibiotics
Kirby-Bauer method
Disks soaked with () are placed on agar plates inoculated with lawn of ()
() diffuses from disk into agar forming ()
Observe () zones around disk
Measure size of () zone to determine ()
Kirby-Bauer method
Disks soaked with (specific drugs) are placed on agar plates inoculated with lawn of ( test microbe)
(Drug) diffuses from disk into agar forming (concentration gradient)
Observe (cleared) zones around disk
Measure size of (clear) zone to determine (MIC)
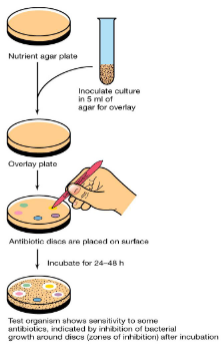
After incubating agar plates in Kirby-Bauer test you measure diameter of () around disks and compare results with () values to see if antibiotic concentration is low enough to be clinically useful
After incubating agar plates in Kirby-Bauer test you measure diameter of (zone of inhibition) around disks and compare results with (MIC) values to see if antibiotic concentration is low enough to be clinically useful
As long as the concentration of drug in tissue or blood is higher than () the drug will be effective
As long as the concentration of drug in tissue or blood is higher than (MIC) the drug will be effective
Gram negative bacteria has a () layer with () while gram positive bacteria has a () layer with ()
Gram negative bacteria has a (thin) layer with (direct cross linking) while gram positive bacteria has a (thick) layer with (peptide interbridge)
Penicillin is a () antibiotic and is a () of crosslink ()
() is an important in this compound as well as other compounds ending in cillin
Penicillin is a (cell wall) antibiotic and is a (competitive inhibitor) of crosslink (transpeptidation)
(Beta lactam) is an important in this compound as well as other compounds ending in cillin
Other cell wall synthesis inhibitors include () which binds ends of peptides, () blocks formation of peptide for crosslink, and () blocks movement across membrane
Other cell wall synthesis inhibitors include (vancomycin) which binds ends of peptides, (cycloserine) blocks formation of peptide for crosslink, and (bacitracin) blocks movement across membrane
() are derived from Cysteine and Valine but have a different () which allows the drug to bind to transpeptidase and transglycosylase, halting cell wall synthesis
(Beta lactams) are derived from Cysteine and Valine but have a different (R group) which allows the drug to bind to transpeptidase and transglycosylase, halting cell wall synthesis
() are enzymes that cleaves Beta lactam ring of Beta lactam
The enzyme is transported out of cell into () of gram positive bacteria or () of gram negative bacteria
(Beta lactamases) are enzymes that cleaves Beta lactam ring of Beta lactam
The enzyme is transported out of cell into (surrounding media) of gram positive bacteria or (periplasm) of gram negative bacteria
() or nalidixic acid is an antibiotic that interferes with () by blocking bacterial DNA gyrase it prevents ()
(Quinolines) or nalidixic acid is an antibiotic that interferes with (DNA synthesis/integrity) by blocking bacterial DNA gyrase it prevents (DNA replication)
() are antibiotics that interfere with bacterial metabolism
(Sulfa drugs/anti-metabolites) are antibiotics that interfere with bacterial metabolism
Most bactericidal are () inhibitors that inhibit transcription like rifampicin which blocks bacterial () and actinomycin D which binds DNA and inhibits () movement
Most bactericidal are (RNA synthesis) inhibitors that inhibit transcription like rifampicin which blocks bacterial (RNA polymerase) and actinomycin D which binds DNA and inhibits (polymerase) movement
Ribosomal inhibitor antibiotics include Macrolides, chloramphenicols, and lincosamides like erythromycin and azithromycin bind large () blocking transfer of ()
Ribosomal inhibitor antibiotics include Macrolides, chloramphenicols, lincosamides like erythromycin and azithromycin bind large (subunit) blocking transfer of (peptides)
Ribosomal inhibitor antibiotics include Aminoglycosides like streptomycin prevent () and () subunits from binding each other
Aminoglycosides like streptomycin prevent (30s) and (50s) subunits from binding each other
Ribosomal inhibitor antibiotics include Tetracyclines which binds () blocking binding of ()
Ribosomal inhibitor antibiotics include Tetracyclines which binds (small subunit) blocking binding of (aminoacyl tRNA)
Some drugs become incorporated into the cell () and damage it causing disruption of () by making holes in the (), killing the cell
() forms a cation channel, () leaks and cell can’t maintain ()
Some drugs become incorporated into the cell (membrane) and damage it causing disruption of (cytoplasmic membrane) by making holes in the (cell membrane), killing the cell
(Gramicidin) forms a cation channel, (H+) leaks and cell can’t maintain (proton motive force)
Methods of Antibiotic Resistance
Destroy () before it gets into cell
Add () that inactivate antibiotic
Modify () so antibiotic no longer () to it
Use () pump to move antibiotic () of cell
Methods of Antibiotic Resistance
Destroy (antibiotic) before it gets into cell
Add (modifying group) that inactivate antibiotic
Modify (target) so antibiotic no longer (binds) to it
Use (efflux) pump to move antibiotic (out) of cell
Many bacteria make () to destroy the antibiotic making the bacteria resistant and lowering penicillin concentration to protect nearby cells
Bacteria can create enzymes using acetylation, phosphorylation, and adenylation that modify () antibiotics that change the antibiotic to no longer interfere with ()
Many bacteria make (Beta lactamase) to destroy the antibiotic making the bacteria resistant and lowering penicillin concentration to protect nearby cells
Bacteria can create enzymes using acetylation, phosphorylation, and adenylation that modify (aminoglycoside) antibiotics that change the antibiotic to no longer interfere with (ribosomes)
The most common streptomycin resistance mechanism is modifying () or () of penicillin
() pumps drug out of cell and often confers resistance to several different classes of antibiotics simultaneously
The most common streptomycin resistance mechanism is modifying (target enzyme) or (ribosome) of penicillin
(Multidrug resistance transporter) pumps drug out of cell and often confers resistance to several different classes of antibiotics simultaneously
Preventing emergence of antibiotic resistance:
Give antibiotics in () concentrations
Give () drugs at the same time
() prescriptions
Use antibiotics ()
Preventing emergence of antibiotic resistance:
Give antibiotics in (high) concentrations
Give (2 or more) drugs at the same time
(Finish) prescriptions
Use antibiotics (only when necessary)
Strategies to prevent or treat drug resistant pathogens
() antibiotic
Use () antibiotic
Use another drug to () enzymes
() is used in combination with penicillin and binds to ()
Strategies to prevent or treat drug resistant pathogens
(Chemically alter) antibiotic
Use (more than 1) antibiotic
Use another drug to (inactivate resistance) enzymes
(Clavulanic acid) is used in combination with penicillin and binds to (Beta lactamases)
Many bacteria have () from trying to protect itself from its own antibiotics as well as against others in its community
Many bacteria have (natural antibiotic resistance) from trying to protect itself from its own antibiotics as well as against others in its community
() can come from incomplete/over use of antibiotics which leads to () on bacteria, () which are very rare () mutations, and () which is getting resistance genes from another through () carrying multiple resistance genes or () like transposons and integrons
(Acquired resistance) can come from incomplete/over use of antibiotics which leads to (increased selective pressure) on bacteria, (antimicrobial resistance mutations) which are very rare (spontaneous) mutations, and (horizontal gene transfer) which is getting resistance genes from another through (R plasmid) carrying multiple resistance genes or (mobile genetic elements) like transposons and integrons
() inhibits viral uncoating and prevents entry of influenza virus into host cell
() blocks neuraminidase and prevents release of mature viruses
(Amantadine) inhibits viral uncoating and prevents entry of influenza virus into host cell
(Zanamivir) blocks neuraminidase and prevents release of mature viruses
Paxlovid uses () to block catalytic site of SARS CoV 2’s main () which prevents protein processing and () to slow liver’s degradation of ()
Paxlovid uses (Nirmatrelvir) to block catalytic site of SARS CoV 2’s main (protease) which prevents protein processing and (Ritonavir) to slow liver’s degradation of (Nirmatrelvir)
E.coli K-12 and Saccharomyces spp. are examples of risk group () organisms where work can be performed on () and personal protective equipment worn ()
E.coli K-12 and Saccharomyces spp. are examples of risk group (1) organisms where work can be performed on (open lab bench/table) and personal protective equipment worn (as needed)
Diarrheagenic E. coli, Samonella spp., and Zika virus are examples of risk group () organisms where personal protective equipment () worn and () is used to store items
Diarrheagenic E. coli, Samonella spp., and Zika virus are examples of risk group (2) organisms where personal protective equipment (is) worn and (biological safety cabinet) is used to store items
SARS and mycobacterium tuberculosis are examples of risk group () organisms where personal protective equipment () worn, () may be used, () are used, and the entrance to lab is through 2 sets of self-closing and locking doors
SARS and mycobacterium tuberculosis are examples of risk group (3) organisms where personal protective equipment (is) worn, (respirators) may be used, (biological safety cabinets) are used, and the entrance to lab is through 2 sets of self-closing and locking doors
Ebola and Hanta virus are examples of risk group () organisms where (), (), () suits are worn and treatments may not be available as there are only a few facilities in the US
Ebola and Hanta virus are examples of risk group (4) organisms where (full body), (air-supplied), (positive pressure) suits are worn and treatments may not be available as there are only a few facilities in the US
Pathogen identification is crucial for () treatment, () issues and track () of disease
Pathogen identification is crucial for (appropriate) treatment, (downstream) issues and track (spread) of disease
Identifying pathogen on () media removes non-pathogenic strains and the ability to grow on medium helps identification while on () media different strains grow but differ in indicator color
Identifying pathogen on (selective) media removes non-pathogenic strains and the ability to grow on medium helps identification while on (differential) media different strains grow but differ in indicator color
() strip technology identifies pathogens by metabolism and resulting reactions causes () that indicate positive or negative reactions
It includes several biochemical tests in 1 strip and each well contains chemicals needed for different biochemical test
(Analytical profile index API) strip technology identifies pathogens by metabolism and resulting reactions causes () that indicate positive or negative reactions
It includes several biochemical tests in 1 strip and each well contains chemicals needed for different biochemical test
() amplifies small fragments of DNA and allows detection of tiny numbers of bacteria
(PCR) amplifies small fragments of DNA and allows detection of tiny numbers of bacteria
() can be used quantify microbial RNAs using reverse transcriptase to generate complementary DNA from RNA templates
(Quantitative real time PCR) can be used quantify microbial RNAs using reverse transcriptase to generate complementary DNA from RNA templates
() can detect antigens or antibodies present in nanogram and picogram quantities
() from virus attached to wells, () binds to (), () have enzymes attached and the overall enzyme reaction causes color change indicating presence of ()
(Enzyme linked immunosorbent assay ELISA) can detect antigens or antibodies present in nanogram and picogram quantities
(Antigen) from virus attached to wells, (antibody) binds to (antigen), (secondary antibodies) have enzymes attached and the overall enzyme reaction causes color change indicating presence of (antivirus antibodies)
() are designed to be used directly at site of patient care and rely on () to give results
An example would be the at home COV kits
(Point of care laboratory tests) are designed to be used directly at site of patient care and rely on (immunochromatography) to give results
An example would be the at home COV kits
Food () refers to microbial changes that render product unfit for consumption while food () refers to presence of human pathogens
Food (spoilage) refers to microbial changes that render product unfit for consumption while food (contamination/poisoning) refers to presence of human pathogens