Lecture 3: Eukaryotic transcription and RNA processing/degradation
1/102
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
103 Terms
What does RNA pol I transcribe?
transcribes rRNA genes (5.8s, 18s, 28s rRNA). only one 1 transcript but responsible for 80% of total RNA in a cell
What does RNA pol II transcribe
transcribes protein coding genes, lncRNA, snoRNA, miRNA, siRNA, and most snRNA (over
What does RNA pol III transcribe
transcribes tRNA, 5s rRNA, some snRNA, and other small RNA
Pol II has — subunits. Most of these subunits share some sort of function that is similar to —--
12; bacterial RNA pol
Rpb1
subunit of RNA pol II. has a specialized C-terminal domain (CTD) which is phosphorylated in certain spots at certain points of transcription.
The structure of the —- of the RNA polymerase is very similar between eukaryotes and bacteria
core enzyme
Pol II requires a number of —--- to initiate transcription
basal transcription factors
Five essential/basal transcription factors for RNA pol II
TFIID, TFIIB, TFIIF, TFIIIE, and TFIIH
What subunits make up TFIID?
made of the TBP (Tata binding) subunit (1) and TAF subunits(~11). TBP is similar to the sigma factor in bacteria.
TFIIH
one subunit has helicase activity. Another is a kinase. Responsible for unwinding DNA at transcription start point and phosphorylation of ser5 of RNA polymerase CTD (on the rpb subunit). This releases RNA polymerase from the promoter to start transcription
What types of eukaryotic promoters are there?
core promoter, proximal promoter, and distal promoter(enhancer)
Promoters may be found where in relation to the transcription start site in eukaryotes?
upstream or downstream
Core promoter
aka minimal promoter. +/- ~40 bp around transcription start site
Proximal promoter
200-400 bp from transcription start site
Distal promoter(enhancer)
may be kilobases from the transcription start site. Can act as a promoter for multiple genes.
Most eukaryotic genes have what conserved sequence in a promoter?
TATA box
In eukaryotes, different genes use a different combination of —--/—- —-- elements
core/proximal promoter
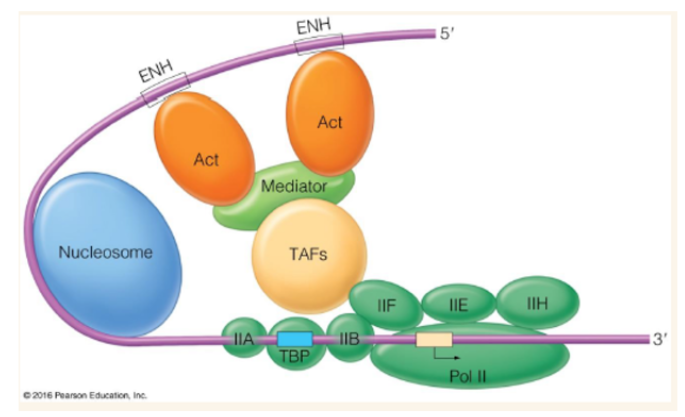
Activator proteins
bind to enhancer (distal promoters) and causes DNA looping to contact activators and basal transcription factors through mediator
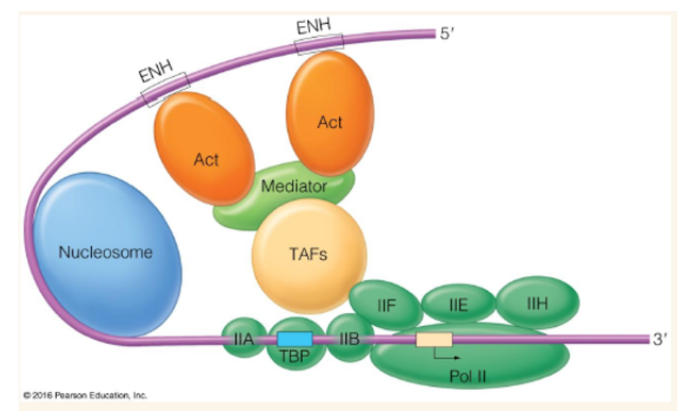
Mediator
huge multiprotein complex that causes DNA looping which brings in contact activators attached to enhancer and basal transcription factors attached to TSS (transcription start site)
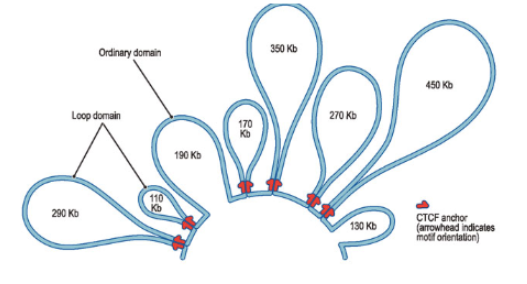
—- define domains of enhancer and promoter DNA that may interact with each other
large DNA loops
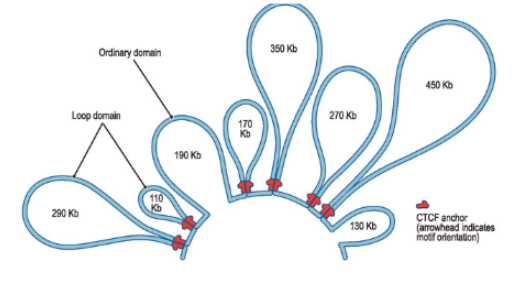
Insulators
establish chromatin boundaries between enhancer/promoter loops (found at the base of large DNA loops)
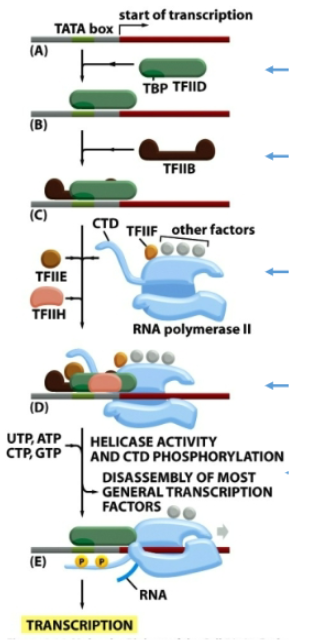
Series of events of transcription initiation by polI
TBP of TFIID binds to TATA box. TFIIB is then recruited to TBP-TATA-complex. This promotes recruitment of RNA pol II with TFIIF to the promoter. Finally TFIIE and TFIIH join the complex (pre-initiation complex is complete). TFIIH helicase activity unwinds dsDNA, it also phosphorylates C-terminal domain (CTD) of Rpb1 which stimulates release of mediator. Most general TFs are released and transcription starts
First region unwound by TFIIH
TATA box
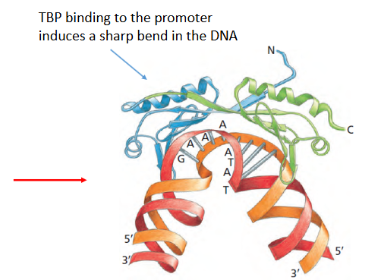
TBP binding induces what in DNA?
a sharp bend
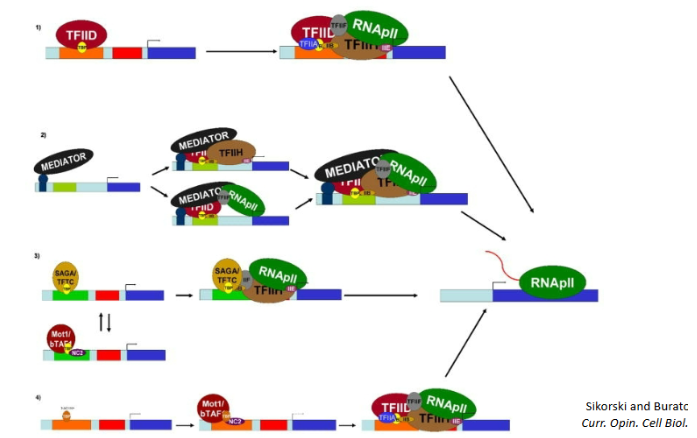
t/f transcription may only be initiated by TFIID TBP binding to TATA box
false, there are many ways for transcription to begin
GRO-seq (global run-on sequencing)
similar to RNA seq. assay to find which RNAs are transcribed the most in a cell. Assay for RNA pol that are actively transcribing and give strand-specific info. Isolate nuclei or permeabilize cells and replace UTP with BrUTP. This creates RNA strands with brominated uracil. An antibody is used to isolate brominated RNA strands. Different adaptors are attached to the 5’ and 3’ ends of the RNA and then a primer is attached and reverse transcriptase is used to make cDNA which is sequenced.
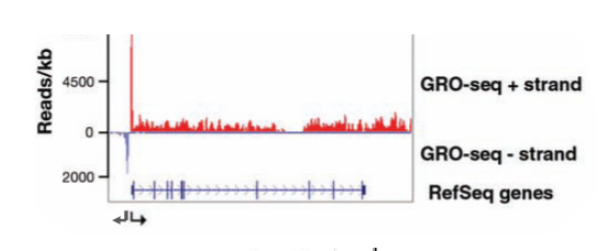
3 surprising results GRO-seq revealed
for many transcripts, polII pauses after starting transcription (promoter-proximal pausing). Transcription in eukaryotes is bidirectional for many if not all genes. Enhancers are also transcribed bidirectionally and produce eRNA which is critical to enhancer function.
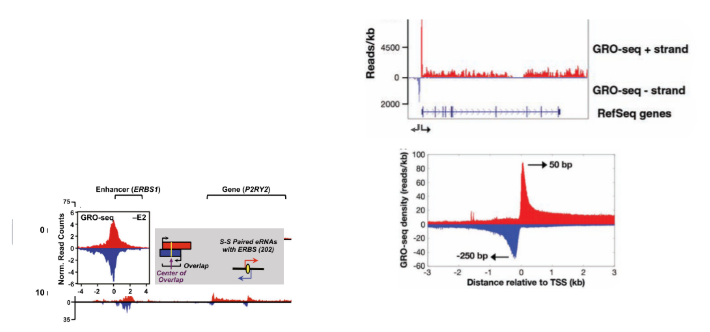
eRNA
RNA produced from enhancer transcription. Critical to function of enhancer
Primary RNA transcript
RNA just after transcription and before modifications
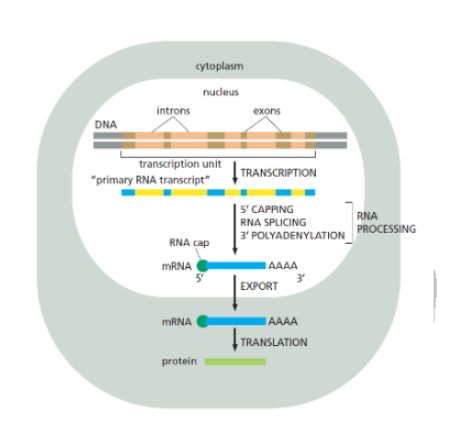
t/f most RNA modifications occur after transcription is finished
false, they occur co-transcriptionally
Modifications required for eukaryotic RNAs
5’ cap, splicing, and polyadenylation
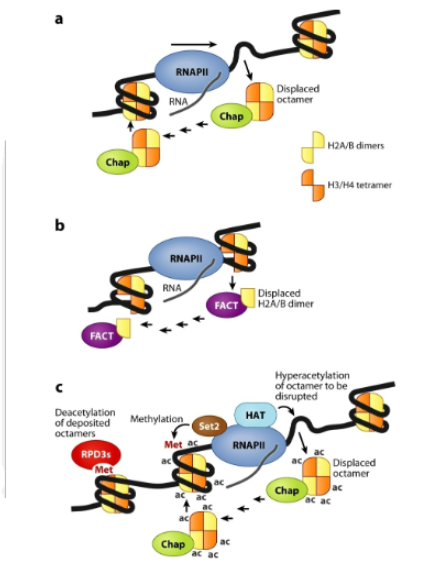
3 ways that eukaryotic RNA polymerases deal with nucleosomes
chaperones displace them and replace them behind the polymerase. Polymerase may transverse through the nucleosome without full displacement. Cotranscriptional modification of histones can help with displacement, replacement and recruitment of other modifiers
t/f RNA pol II usually stops transcription at a polyadenylation signal
false, it usually continues for a long time after the signal
pre-mRNA is cleaved — to — nt after the polyadenylation signal
11-30
polyA polymerase
adds polyA tail to mRNA (up to 300 residues) which affects mRNA stability (longer polyA tail means longer half life of mRNA in eukaryotes)
Longer polyA tail in eukaryotes means what ?
increased mRNA half life
Allosteric model of transcription termination
binding of 3’ processing factors lead to rearrangement of elongation complex and termination
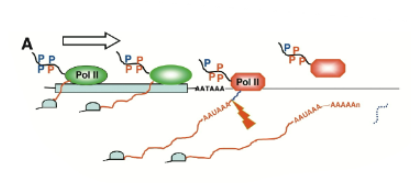
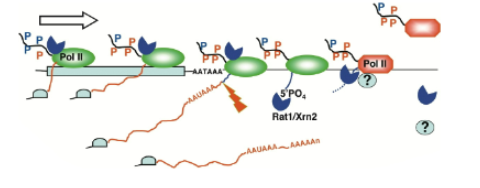
Torpedo model of transcription termination
after cleavage near the poly-adenylaiton sequence, nuclease degrades nascent RNA from 5’ end and catches up with elongation complex to displace pol II from DNA

Combination model of transcription termination
combination of torpedo and allosteric model. Nuclease degrades nascent RNA from 5’ end and causes a rearrangement of the elongation complex and termination whenever it catches up to the elongation complex
RNA polymerase 1 transcribes how many different genes?
1
RNA pol I promoters are —- which means what?
bipartite; upstream of the TSS is two conserved sequence where the polymerase binds
RNA pol III promoters are found where ?
internal (inside the genes that it transcribes)
Alpha amanitin
toxin which causes RNA pols to stop working. RNA pol II is most sensitive to this toxin while pol 1 is insensitive. Pol III is intermediately sensitive.
Where are each of the three RNA pols found in the cell?
RNA II and III are found in the nucleus while pol I is found in the nucleolus
Types of genes transcribed by RNA pol I
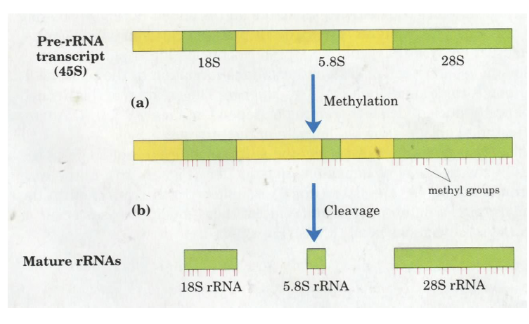
5.8S, 18S, 28S RNA
Types of genes transcribed by RNA pol II
protein-coding genes, lncRNA, snoRNA, miRNA, siRNA, most snRNA
Types of genes transcribed by RNA pol III
tRNA, 5S rRNA, some snRNA, other small RNA
Co-transcriptional modification are facilitated by —-
CTD of polI
What is the sequence of the heptapeptide repeat in the CTD?
YSPTSPS
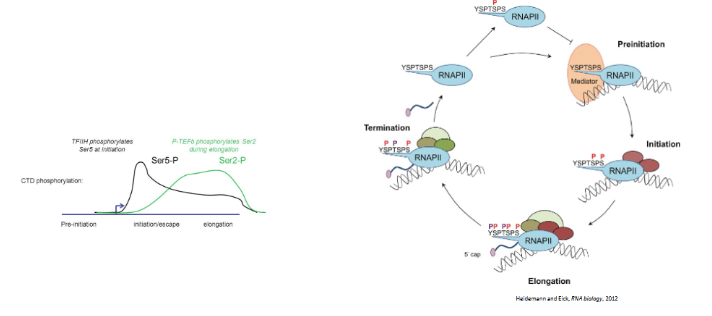
C-terminal domain (CTD)
part of Rpb1 of RNA pol II. contains 15-50+ heptapeptide repeats (YSPTSPS in humans). Depending on how serines are phosphorylated, different factors are attracted to the CTD.
CTD cycle
only serine 5 may be phosphorylated at the beginning of transcription. S5-P also attracts 5’ capping factors. When serine 2 and 5 is phosphorylated, the CTD attracts elongation factors allowing transcription to continue and splicing factors for intron splicing. When only serine 2 is phosphorylated, the CTD attracts termination factors. Serine 7 phosphorylation is important for transcription of snRNAs
Apart from serines, what else can be phosphorylated on the CTD?
Y1 and T4
Why does transcription stop after about 40 bp after initiation?
the RNA pol II must wait for serine II of the CTD to be phosphorylated so that elongation factors may be attracted
1st modification made on RNA while its being synthesized?
5’ cap
—- is added to the 5’ end of all pol II transcripts
7-methylG cap
What does the 5’ cap do for RNA transcripts?
protects the 5’ end from nucleases, helps with translation initiation and facilitation of splicing the 1st intron
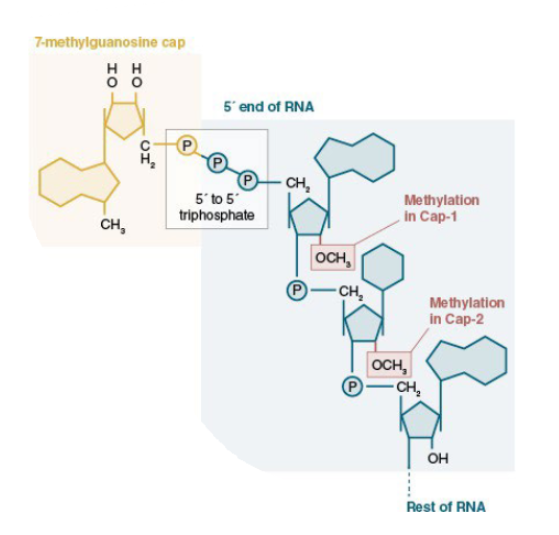
Show what the 5’ cap looks like
5’-5’ phosphodiester linkage between 7mg cap and first nucleotide
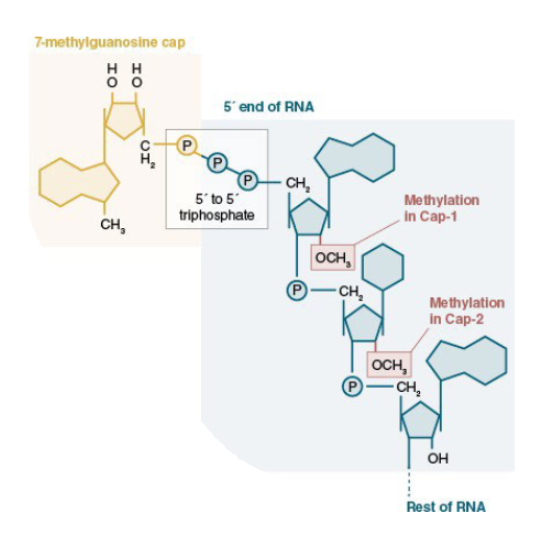
Cap-0 vs cap-1 vs cap-2
cap-0 refers to the 7-methyG cap attached via a phosphodiester linkage to the first nucleotide. The first and/or second nucleotide may be methylated on the 2’ hydroxyl as well (cap-1, cap-2 respectively)
All pol II transcript are polyadenylated at the 3’ except — and —
all snRNAs and smore mRNAs (histones)
How is the poly-A tail produced in eukaryotes
factors recognize the polyadenylation signal on the RNA and cleave off RNA after the signal. polyA-polymerase (PAP) adds a poly-A tail starting from that cleavage site.
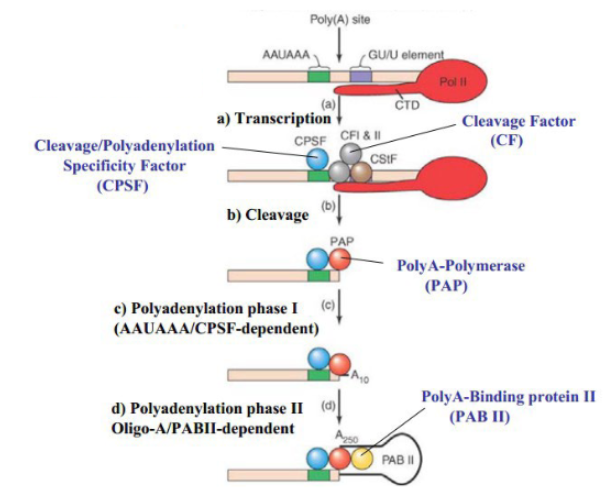
PolyA-binding protein (PAB)
proteins which attach to polyA tails of mRNA and fold it to protect the RNA from 3’ RNAse.
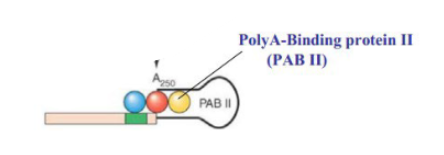
Functions of the poly-A tail in eukaryotes
helps with initiation of translation, coated by PABs (poly-A binding proteins) which protects the 3’ end from nucleases. Facilitates splicing of the last intron
In general a (longer or shorter) poly A tail indicates a more stable RNA in eukaryotes
longer
Both the 7mG cap and polyadenylation carry out what two functions in eukaryotes?
promote translation and increase mRNA stability
A mature RNA transcript is made up of only — sequences
exon
The intron excision process must be precise. Why?
if not precise, a frameshift may be introduced
Spliceosomes do what? What are they composed of?
remove majority of introns; made of 5 snRNA subunits and ~150 proteins
Major spliceosome (snRNA subunits and boundaries)
splices majority of genes in eukaryotes. Subunits are U1,U2,U4,U5, and U6. intron boundaries are GU(5’) and AG(3’).
Minor spliceosome(snRNA subunits and boundaries)
subunits are U11, U12, U4atac, U5, and U6atac. Intron boundaries are Au(5’) and AC(3’)
How are introns marked for splicing
U1 or U11 subunit binds to 5’ splice site
Conserved sequences of DNA identified by a major spliceosome
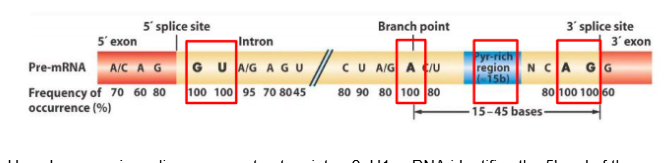
How does a major spliceosome cut out an intron?
U1 snRNA identifies the 5' end of the intron and the U2 snRNA binds to the branch point near the 3’ end. The other snRNA subunits arrive and bend the RNA so that a loop forms. U6 catalyzes cutting and joining of exons ends and a lariat loop is released.
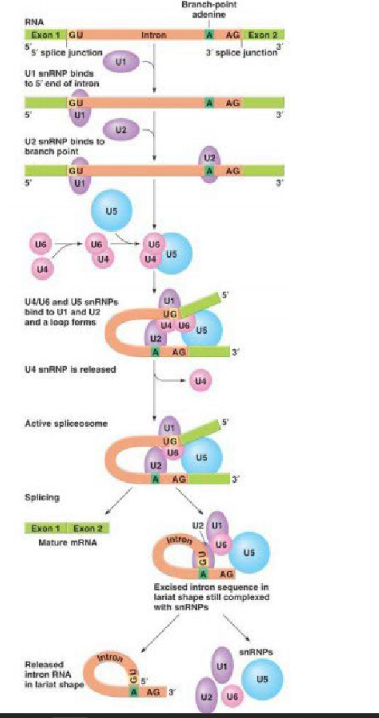
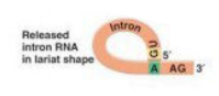
Lariat
circular RNA produced after a spliceosome cuts out an intron. In the case of a major spliceosome, Consists of the 5' splice junction bound to the branch point A
What major spliceosome subunit catalyzes the splicing of introns?
U6 RNA (ribozyme)
Self-splicing introns
introns that are able to splice themself without a spliceosome.
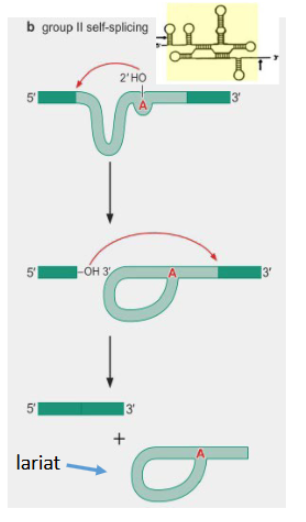
Group II self splicing introns
introns that are able to splice themself without a spliceosome. Still has a branch point A which allows for release of a lariat after splicing.
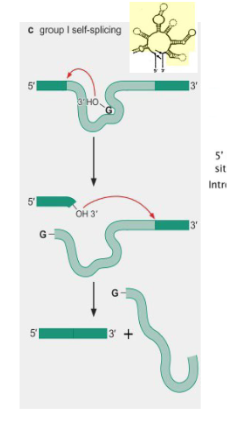
Group I self splicing introns
introns that are able to splice themself without a spliceosome. Does not have a branch point A, does not release a lariat after splicing.
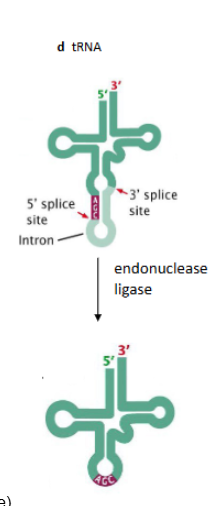
How are tRNAs spliced?
they are spliced via an endonuclease and ligase (not a spliceosome)
Alternative splice forms
different mRNA transcripts may be created from a single gene based on how introns/exons are spliced in that gnee
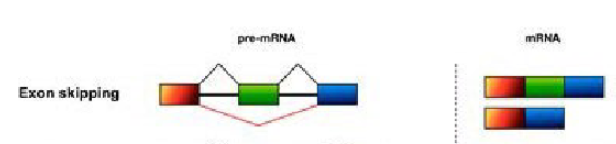
Exon skipping (Alternative splice forms)
An exon may be either included or excluded from the final mRNA product, creating variants that either contain or lack that specific exon.
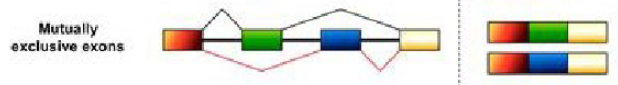
Mutually exclusive zone (Alternative splice forms)
When one of two possible exons is chosen to be included in the mRNA, but not both.
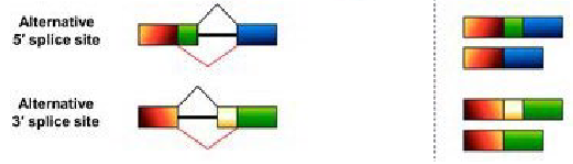
Alternative splice site (3’ or 5’) (Alternative splice forms)
A different 5' splice site is used, changing the length of the downstream exon. A different 3' splice site is used, changing the length of the upstream exon.
Intron retention (Alternative splice forms)
An intron is retained in the mature mRNA instead of being spliced out

Base editing of RNAs
alters RNA sequence and may change protein sequence. More common in tRNA, less common in rRNA, present in mRNA
A to I editing
RNA modification performed by ADARs. Irreversible. Happens in mRNA for synaptic receptors and in processing of some ncRNAs like micro RNAs. changes adenine to inosine.
A to I editing is essential to what?
embryonic development, neuronal development, innate immunity, and processing and maturation of miRNAs
C to U editing
RNA modification performed by APOBECs. Irreversible. Changes cytosine to uracil.
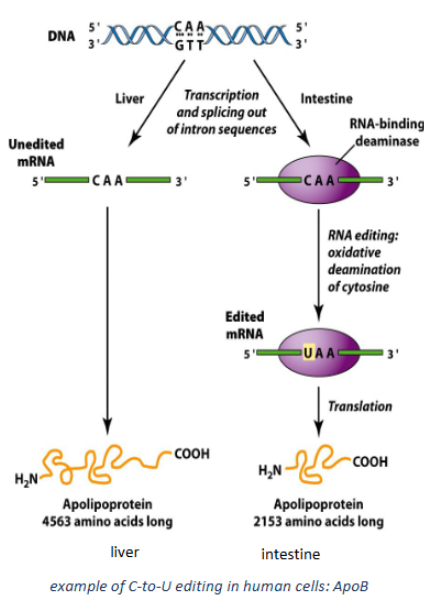
How is C-to-U editing used with ApoB
apolipoprotein B is ,much different in liver vs intestine to to C to U editing creating a stop codon in intestines
mRNA methylation
transient(reversible). Involves methylation of parts of a RNA
Most common mRNA methylation
N6-methyladenosine
mRNA methylation Readers
proteins that recognize and bind methylated mRNA
mRNA methylation Writers
proteins that cause mRNA methylation
mRNA methylation Eraser
reverses mRNA methylation
Different effects of mRNA methylation
increased or decreased mRNA stability, increased or decreased translation, modulation of splicing or others.
How are rRNAs processed?
they are all processed from a single transcript
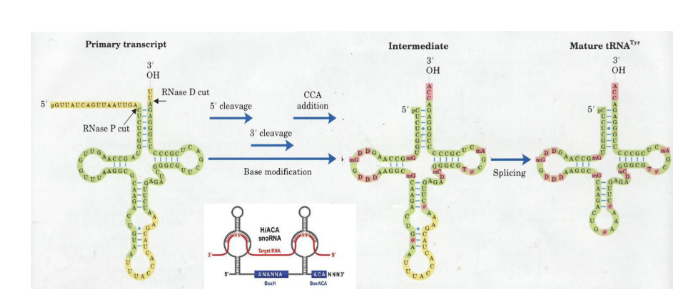
How are tRNAs processed?
processed in part by RNP ribozyme (RNase P). Base modifications are guided by snoRNA (small nucleolar RNA). A single intron is removed via an endonuclease and ligase.
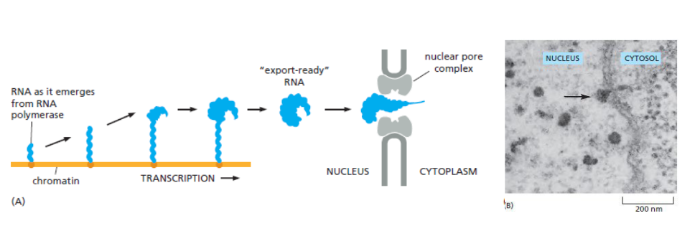
How are mature mRNA exported out of the nucleus?
they are packaged with a number of proteins and are actively transported through the nuclear pore. This process is regulated, so some mRNAs may be transported through a pore more quickly than others.
mRNA surveillance
The 1st round of translation is designed to check for defects in the mRNAs. 3 types of mechanisms
NSD (non-stop decay)
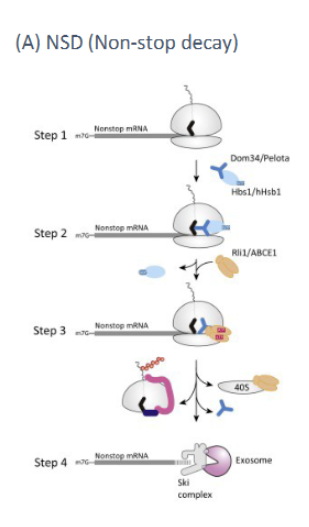
mRNA surveillance mechanism which detects mRNAs with no stop codon. (ie abortive transcription). Ribosomes translate until the end of the mRNA and do not detach due to no stop codon. Proteins recognize this complex, remove the ribosome and an exonuclease chews up the mRNA from the 3’ end
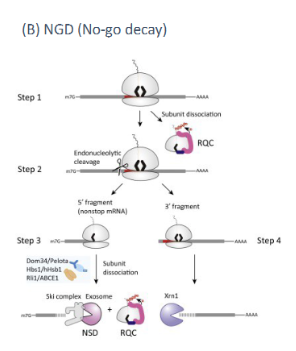
NGD (no-go decay)
mRNA surveillance mechanism which detects major secondary structure issues(usually caused by incorrect intron retention). Detected by stalling of the ribosome at a certain point in translation. Recruits endonucleases to cut out the ribosome. Creates two RNA fragments one with an open 5’ end and another with an open 3’ end that are digested by 5’ and 3’ exonucleases respectively.