BE 359 Ch. 10 Post-Transcriptional Gene Control
1/99
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
100 Terms
RNA
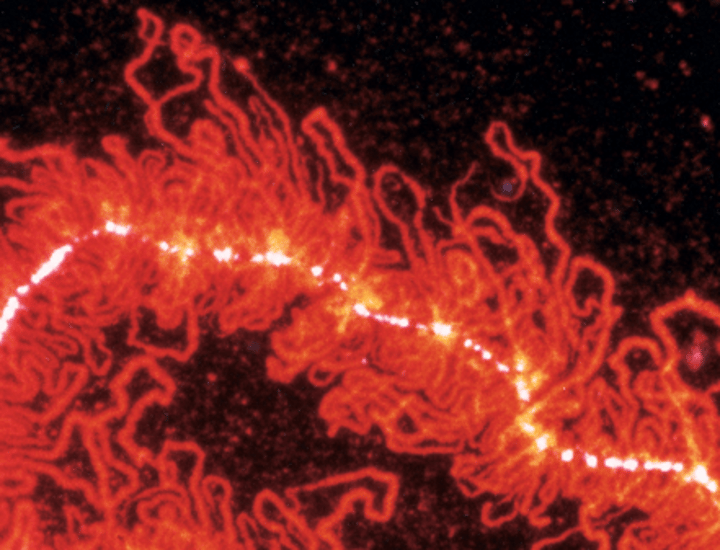
lampbrush chromosome, hnRNP protein associated with nascent _________ transcripts stained red with a monoclonal antibody
pre mRNA
______-_________ is capped, polyadenylated, spliced, and associated with RNPs in the nucleus before export to the cytoplasm
ribonucleoprotein spliceosome
A large _______________ _____________ complex catalyzes two transesterification reactions that join two exons and remove the intron as a lariat structure
splice sites
a network of interactions between SR proteins, snRNPs, and splicing factors forms a cross-exon recognition complex that specifies correct ________ _________
transcriptional regulation, mRNA processing, cytoplasmic regulation
gene expression regulation
RNA polymerase II
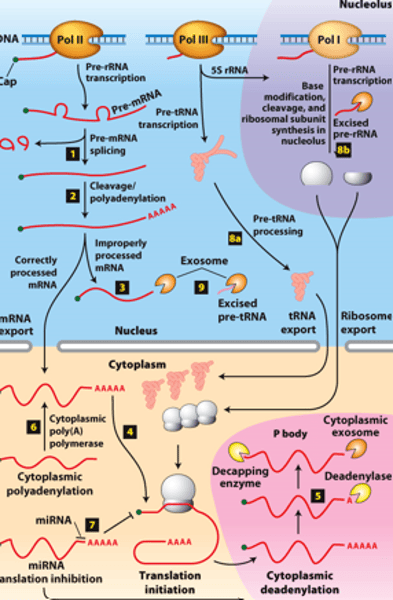
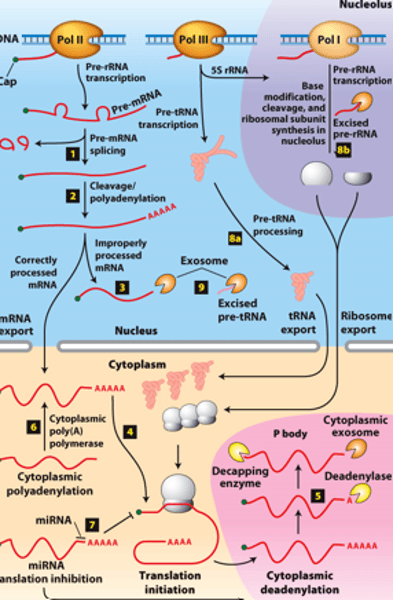
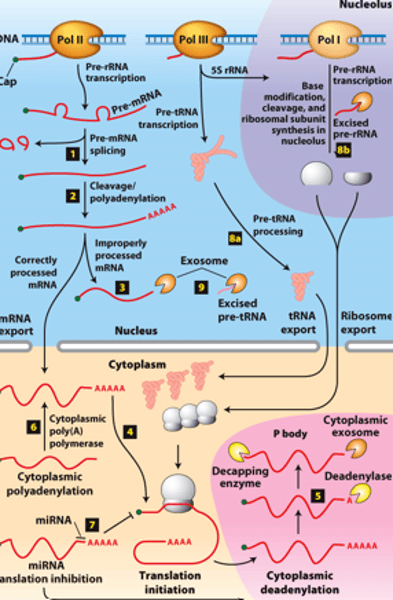
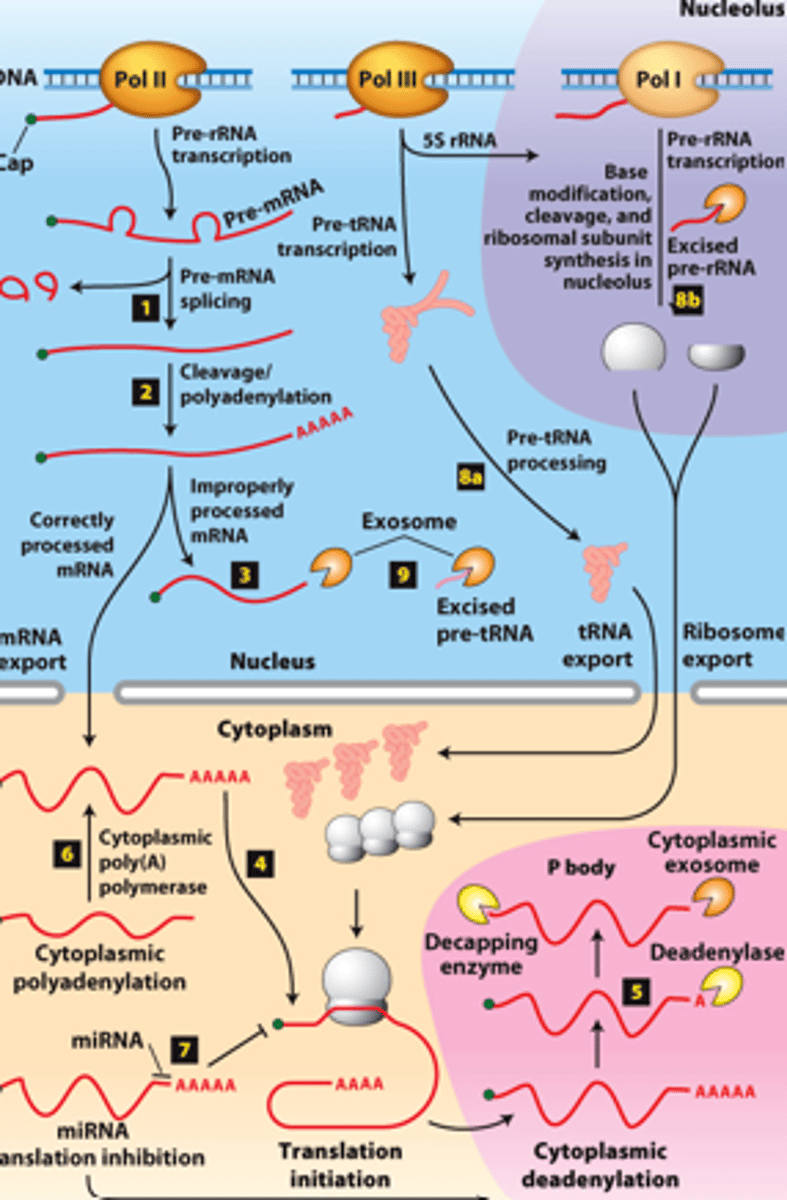
RNA processing:
RNAs from protein-coding genes -
transcribed by ________ ______________ ______
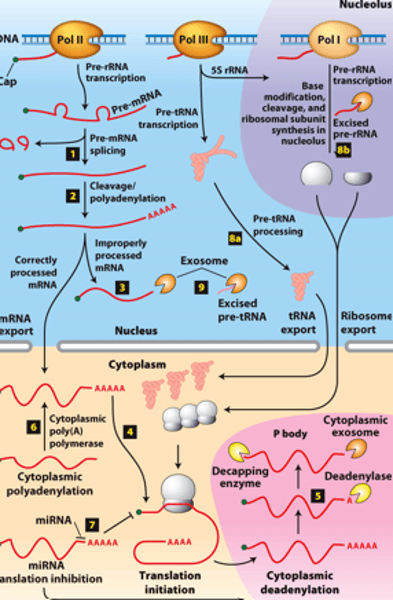
cytoplasm
RNA processing: RNAs from protein-coding genes -processed from primary transcripts in the nucleus before export to the ___________
alternative exons
RNA processing, step 1: use of ____________ ________ during pre-mRNA splicing
poly(A)
RNA processing step 2: use of alternative ____________ sites
exported
RNA processing, step 3:
properly processed mRNAs - _____________ to the cytoplasm
blocked
RNA processing, step 3: improperly processed mRNAs -
___________ from export to the cytoplasm
degraded
RNA processing, step 3: improperly processed mRNAs - ____________ in the exosome complex containing multiple ribonucleases
translation
RNA processing, step 4: translation initiation factors - bind to the 5′ cap cooperatively with poly(A)-binding protein I bound to the poly(A) tail and initiate ____________
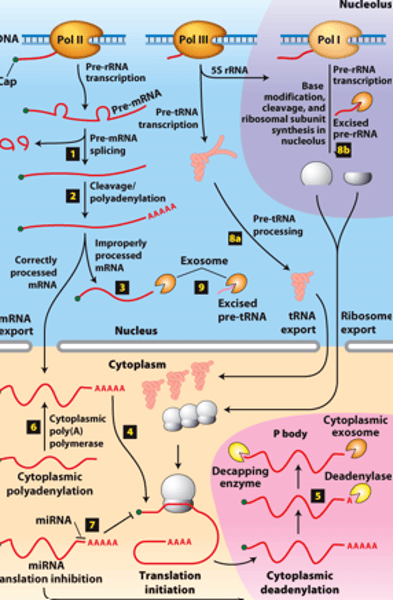
mRNA degraded
RNA processing, step 5: ________ _______________ in cytoplasmic P bodies - translational repression,
deadenylated and decapped by enzymes
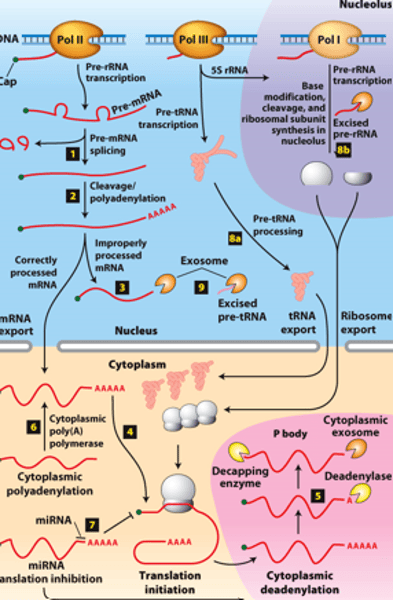
exosomes
RNA processing, step 5: degraded by cytoplasmic _____________
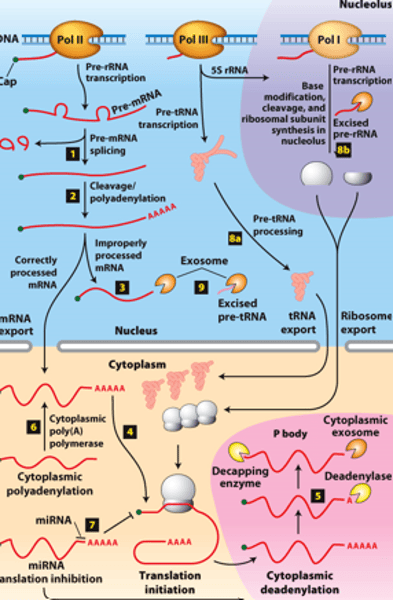
protein translated
RNA processing, step 5: of mRNA degradation rate - regulates mRNA abundance and amount of __________ ______________
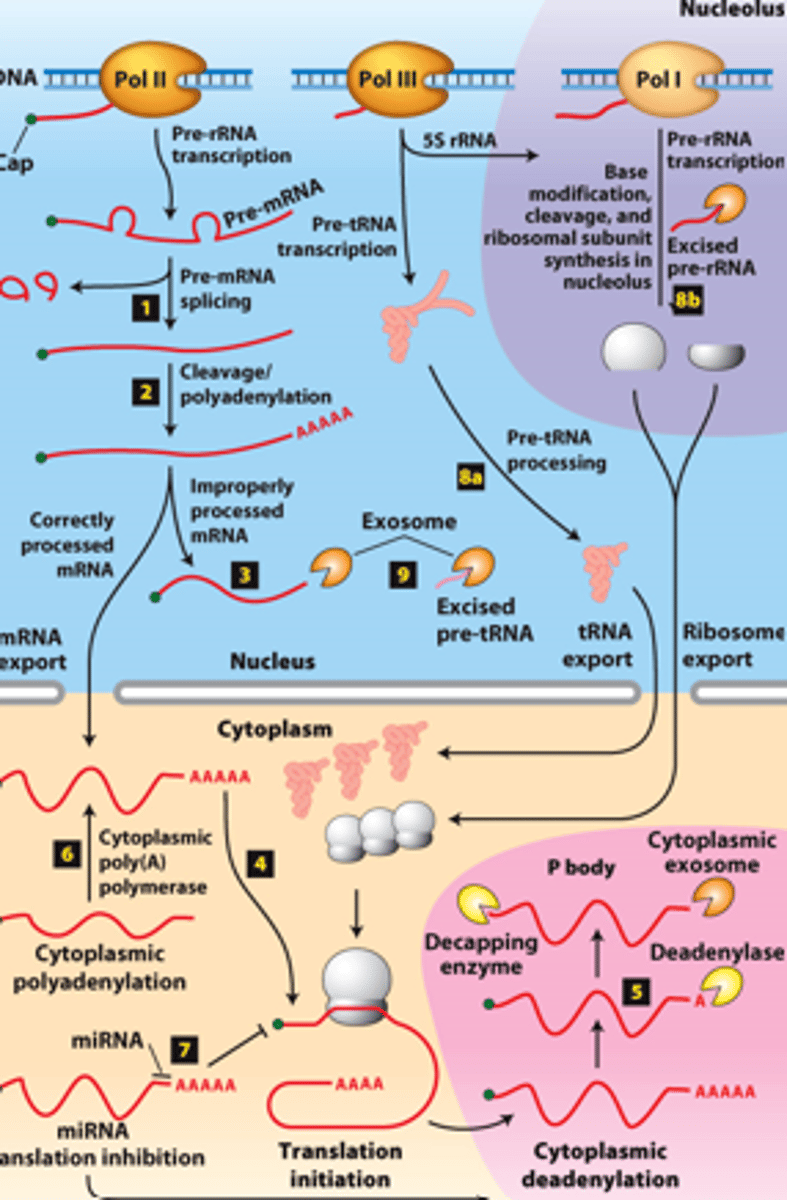
polyA polymerase
RNA processing, step 6 : mRNAs synthesized without long poly(A) tails - translation regulated by controlled synthesis of a long poly(A) tail by a cytoplasmic ___________ ______________
inhibit translation
RNA processing, step 7 : translation regulation by other mechanisms--miRNA (~22-nucleotide RNAs) - ________ _____________ of mRNAs to which they hybridize, usually in the 3′ untranslated region
tRNAs
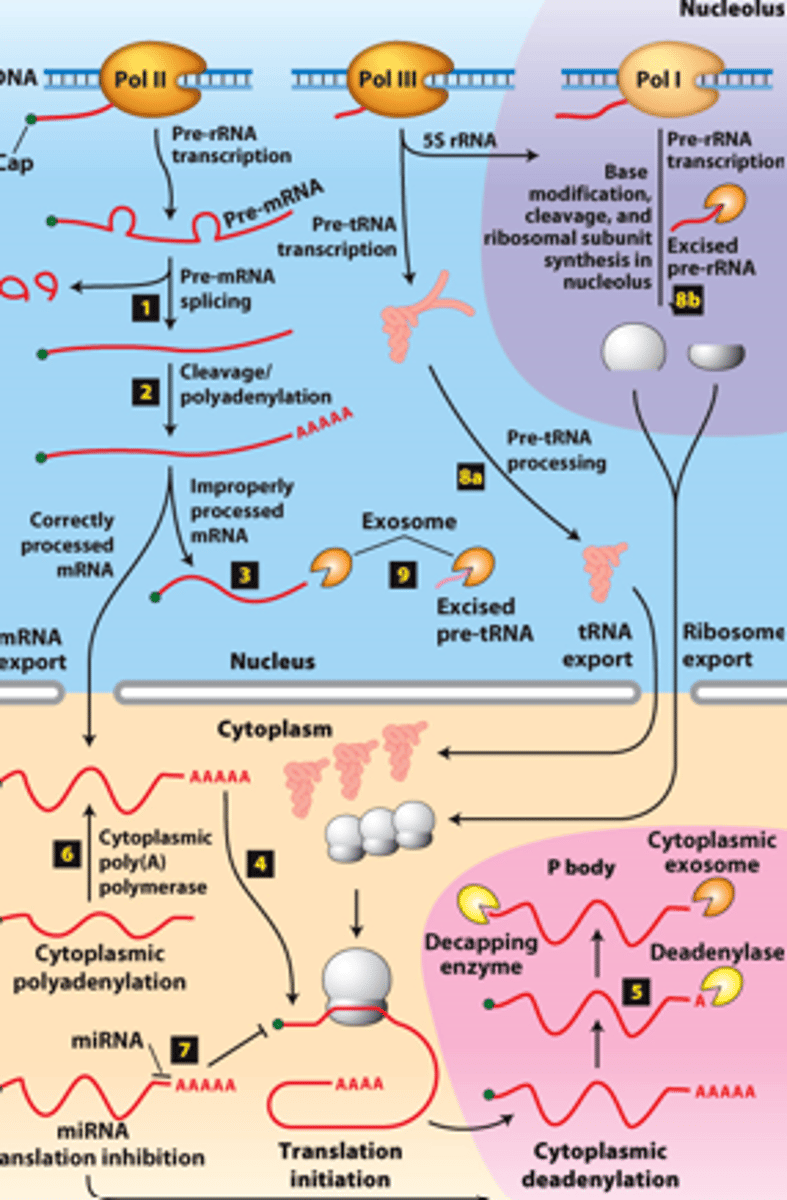
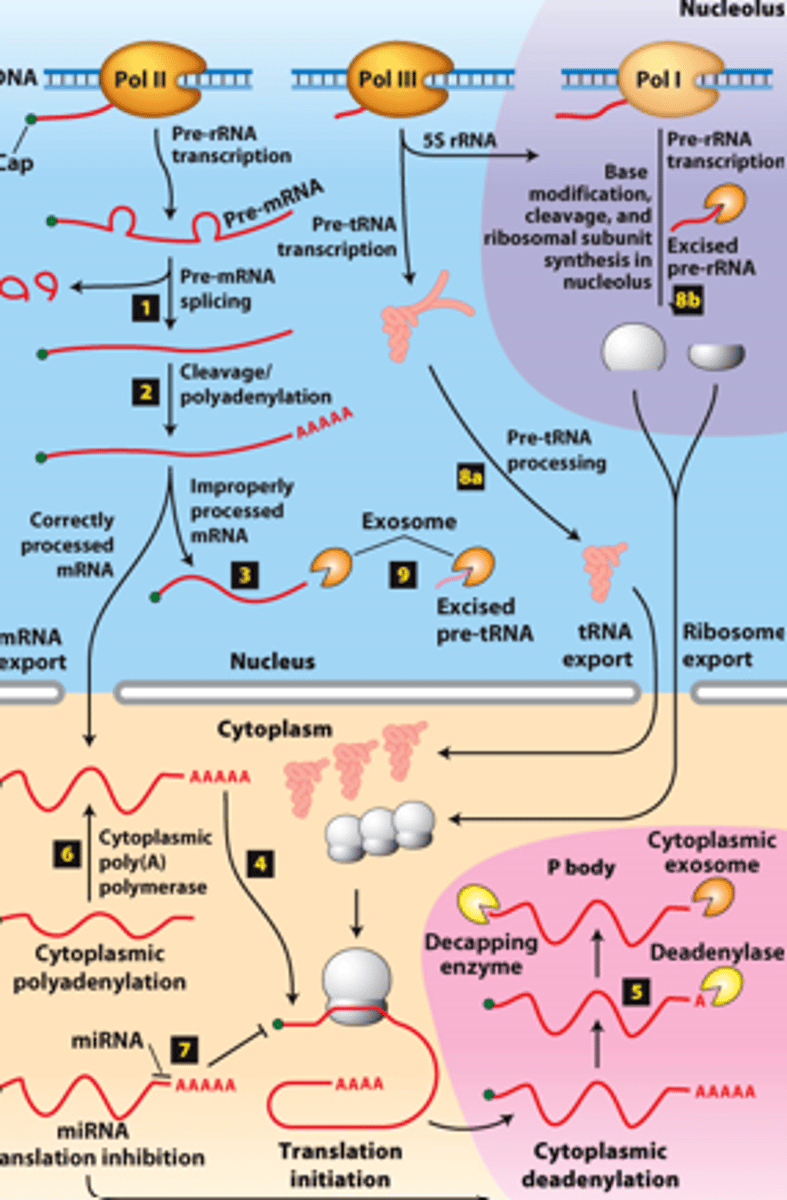
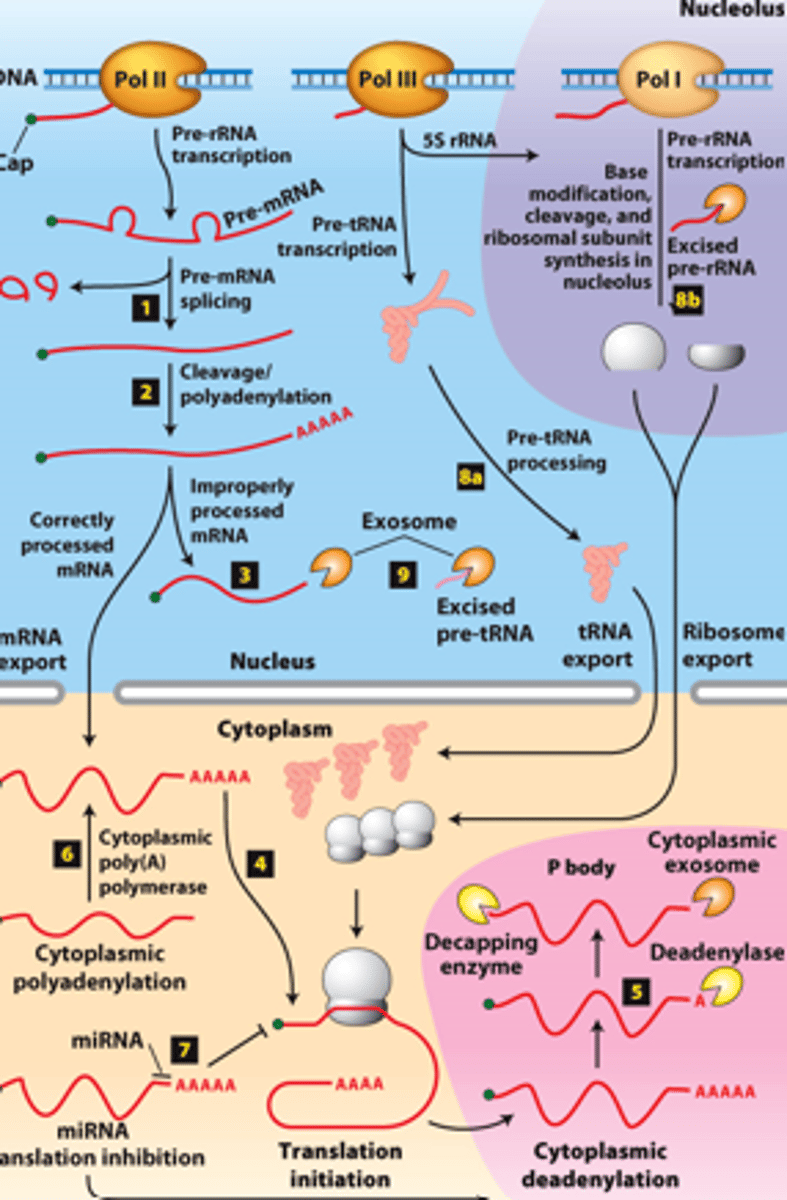
RNA processing, step 8a: __________- transcribed by Pol III and processed in the nucleus
rRNAs
RNA processing, step 8b: __________ - transcribed by Pol I - processed in the nucleolus
mature RNAs
RNA processing, step 9: regions of precursors cleaved from the __________ _______ - degraded by nuclear exosomes
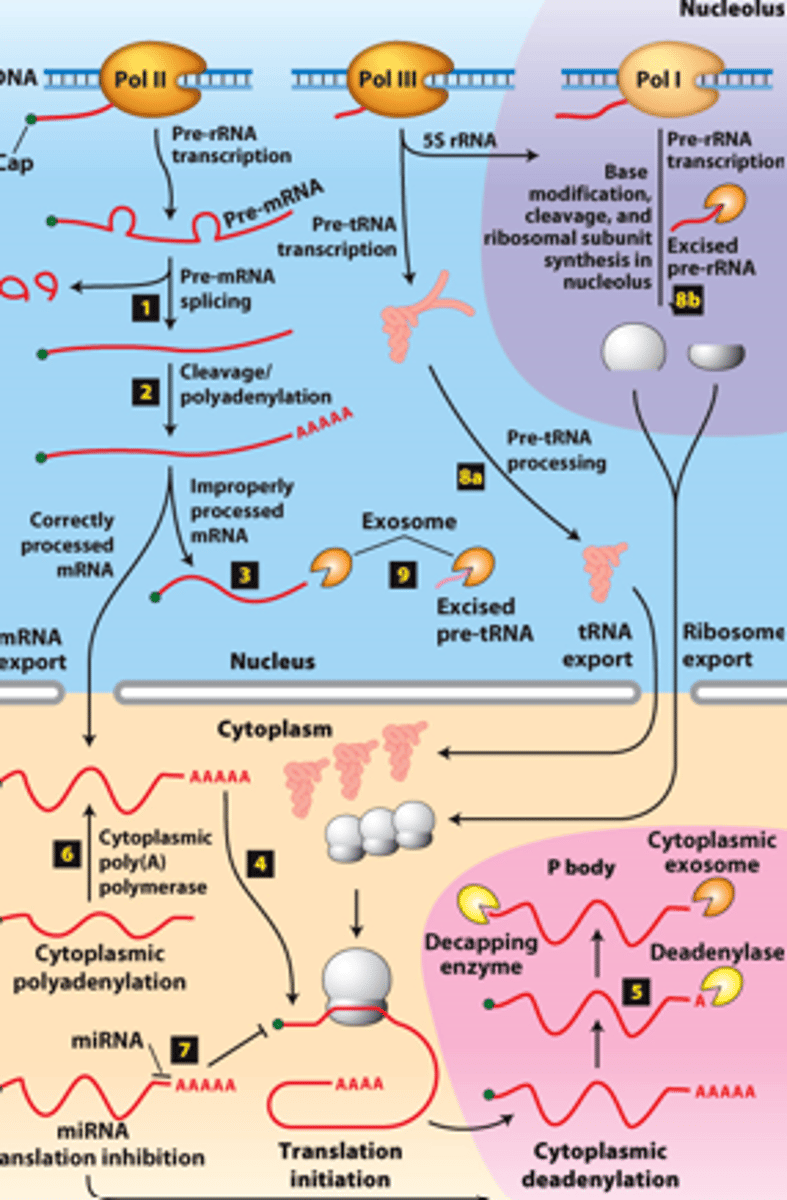
transcription, 5' capping
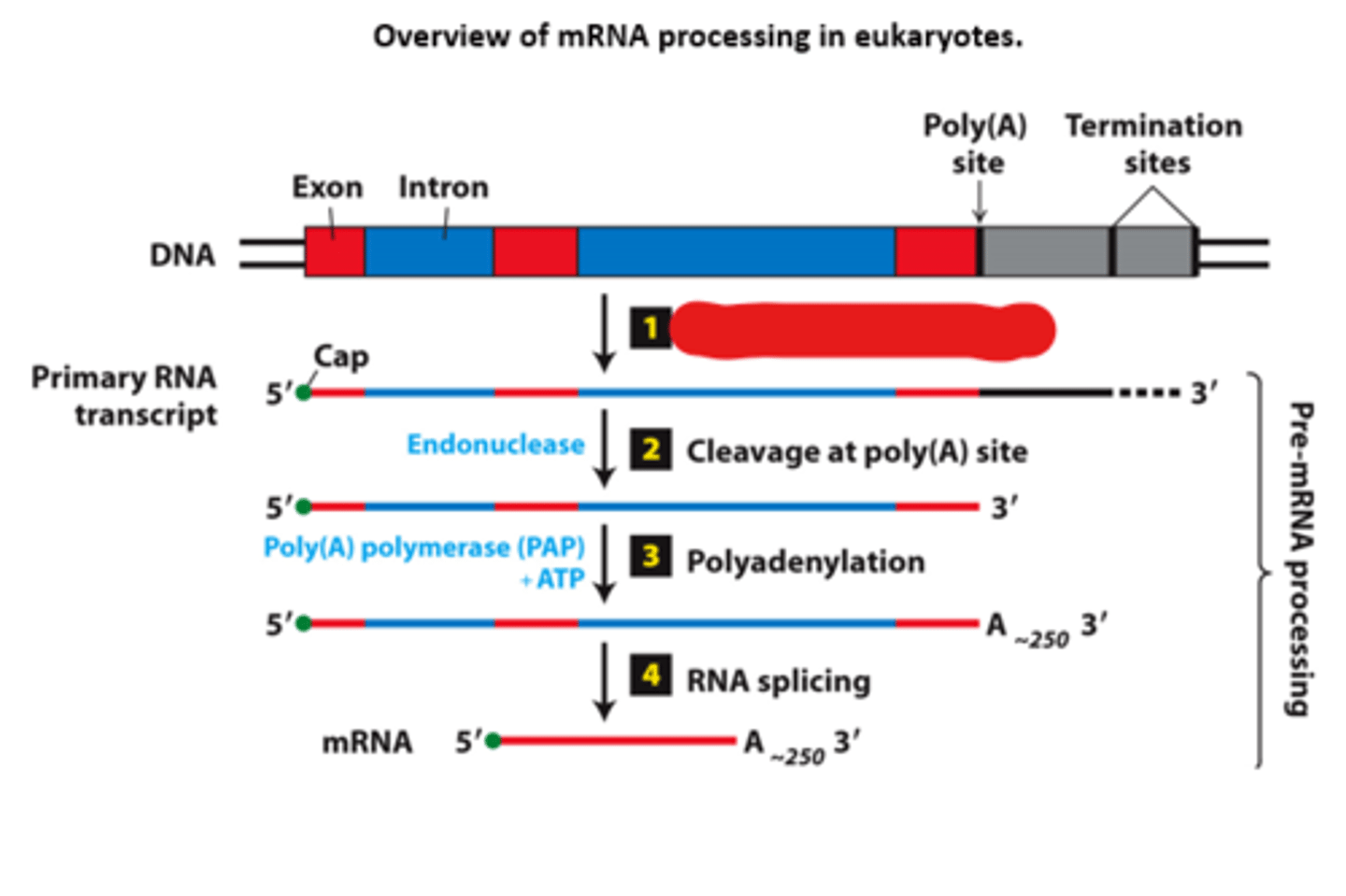
cleavage at polyA site
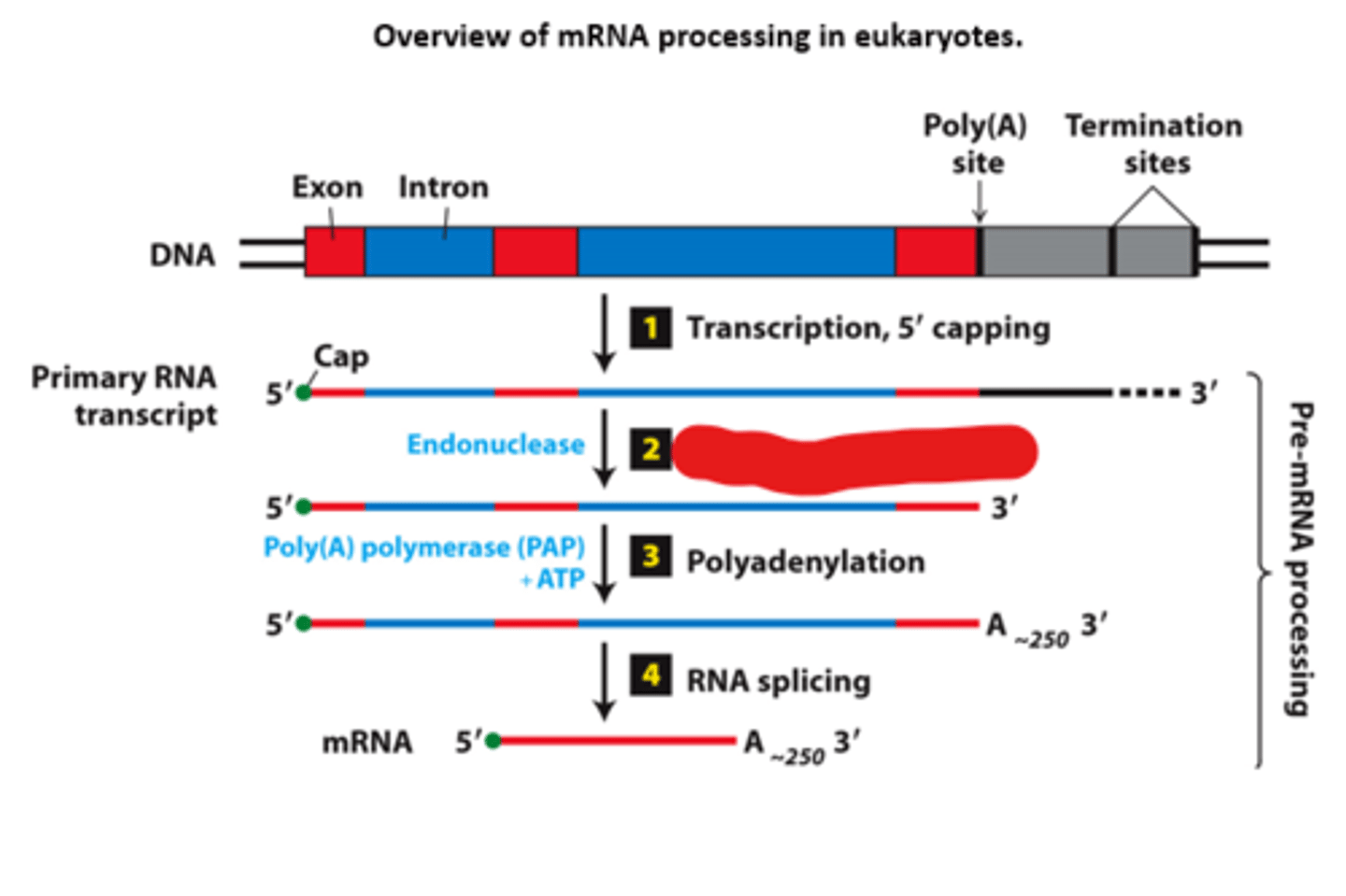
polyadenylation
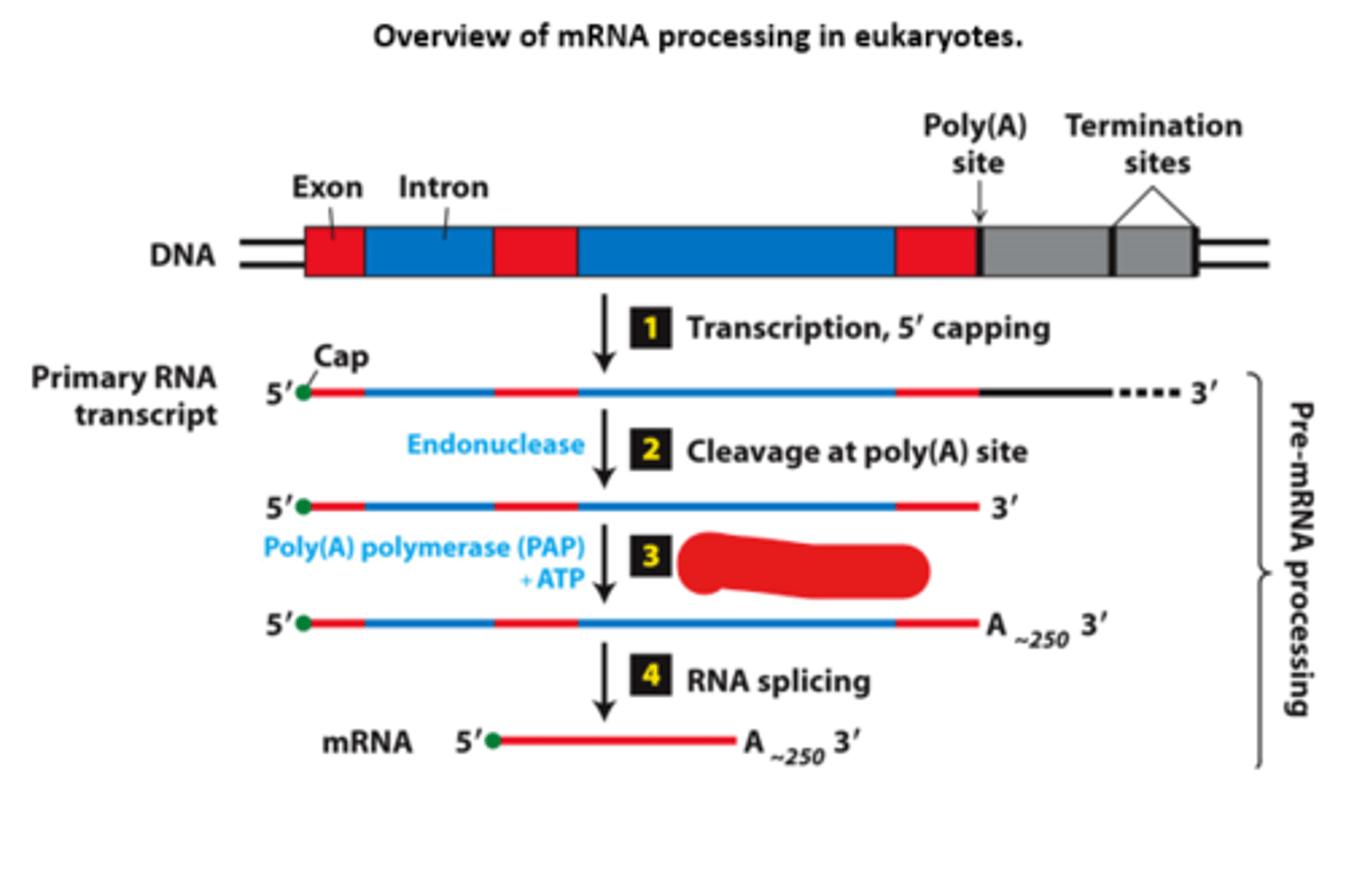
RNA splicing
mRNA
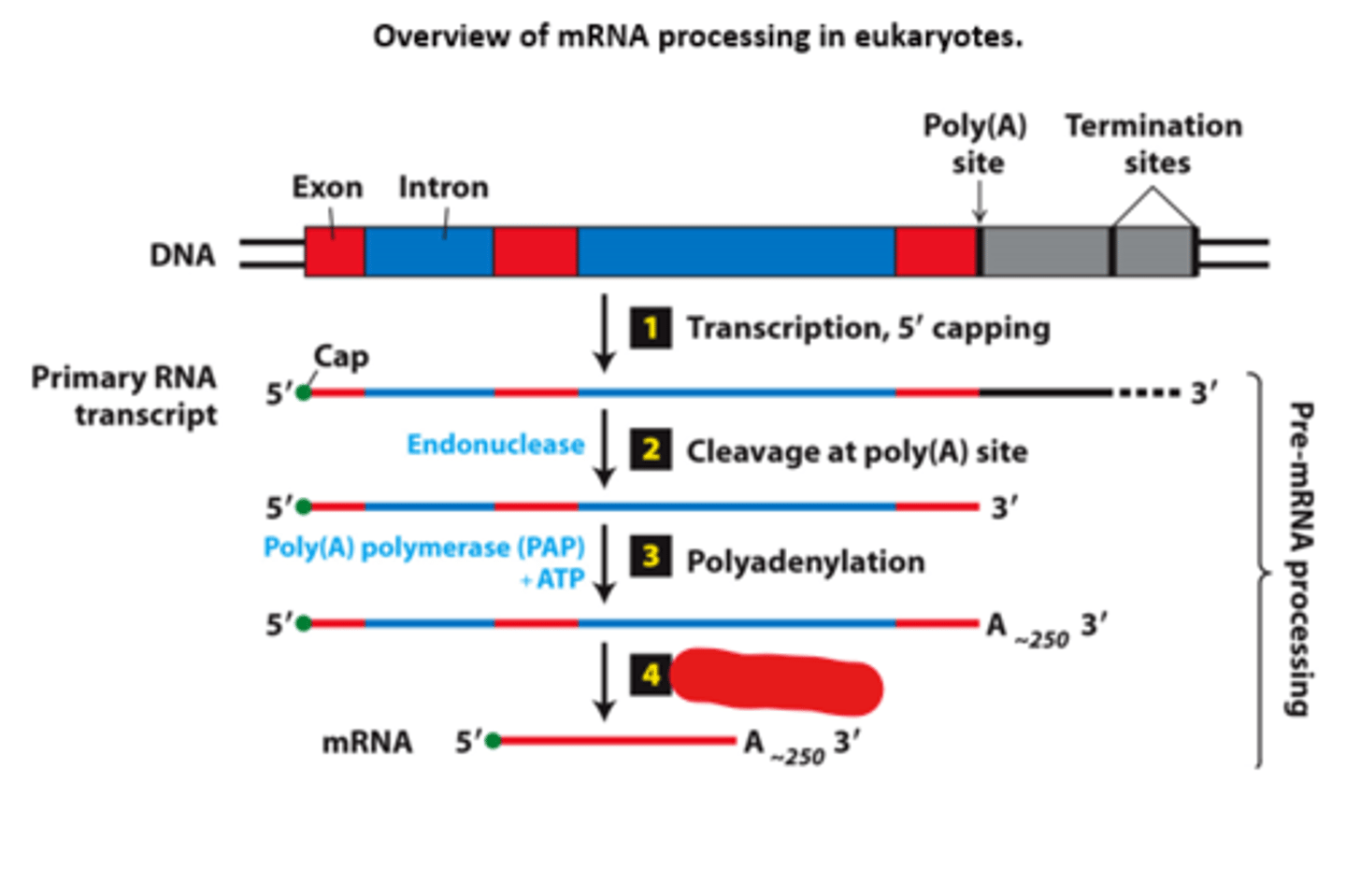
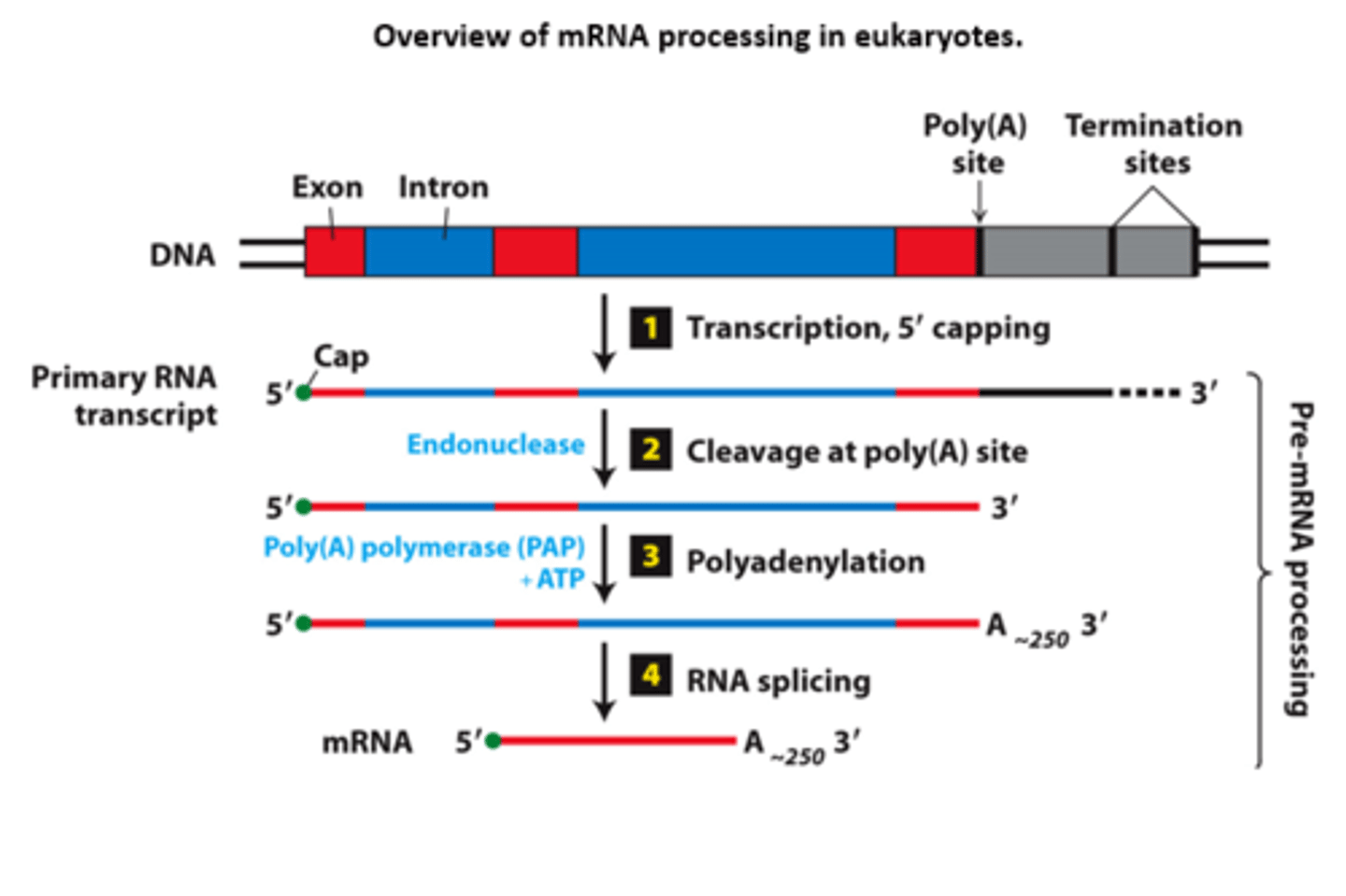
overview of __________ processing in eukaryotes
RNA polymerase II
eukaryotic cells convert an initial ________ ____________ ____ primary transcript into a functional mRNA by three modifications
5' capping, 3' cleavage and polyadenylation, RNA splicing
Eukaryotic cells convert an initial RNA polymerase II primary transcript into a functional mRNA by three modifications
initiates transcription
mRNA processing, step 1: nascent RNA (β-globin RNA) 5′ end capped with 7-methylguanylate (shortly after RNA polymerase II __________ ____________ at the first nucleotide of the first exon of a gene)
termination sites
mRNA processing, step 1: pol II transcription - terminates at any one of multiple ____________ ________ downstream from the poly(A) site in final exon
primary transcript
mRNA processing, step 2: cleavage enzyme cleaves __________ ___________ at the poly(A) site
polyadenylation
mRNA processig, step 3: ________________ enzyme adds a string of adenosine (A) residues (~250 A residues in mammals, ~150 in insects, and ~100 in yeast)
cleavage
mRNA processing, step 4:
short primary transcripts with few introns - splicing follows __________ and polyadenylation (shown)
nascent
mRNA processing, step 4: long transcripts with multiple introns--introns spliced out of the _________ RNA during transcription
mRNA
fully processed messenger RNA with 5' cap, introns removed RNA splicing, and a poly(A) tail
pre-mRNA
an mRNA precursor containing introns and not cleaved at the poly(A) site
hnRNA
heterogeneous nuclear RNAs, these RNAs include pre-mRNAs and RNA-processing intermediates containing one or more introns
snRNA
five small nuclear RNAs that function in the removal of introns from pre-mRNAs by RNA splicing, plus two small nuclear RNAs that substitute for the first two at rare introns
pre-tRNA
a tRNA precursor containing additional transcribed bases at the 5' and 3' ends compared with the mature tRNA, some pre-tRNAs also contain an intron in the anticodon loop
pre-rRNA
the precursor to mature 18S, 5.8S, and 28S ribosomal RNAs, the mature rRNAs are processed from this long precursor RNA molecule by cleavage, removal of bases from the ends of the cleaved products, and modification of specific bases
snoRNA
small nucleolar RNAs, these RNAs baes-pair with complementary regions of the pre-rRNA molecule, directing cleavage of the RNA chain and modification of bases during maturation of the rRNAs
siRNA
short interfering RNAs, around 22 bases long, that are each perfectly complementary to a sequence in an mRNA, together with associated proteins, siRNAs cause cleavage of the "target" RNA, leading to its rapid degradation
miRNA
micro-RNAs, around 22 bases long, that base-pair extensively, but not completely, with mRNAs, especially over bases 2 to 7 at the 5' end of the miRNA (the "seed" sequence), this pairing inhibits translation of the "target" mRNA and targets it for degradation
premRNA splicing
two transesterification reactions at short, conserved sequences
splice
Consensus sequences around _______ sites in vertebrate pre-mRNAs
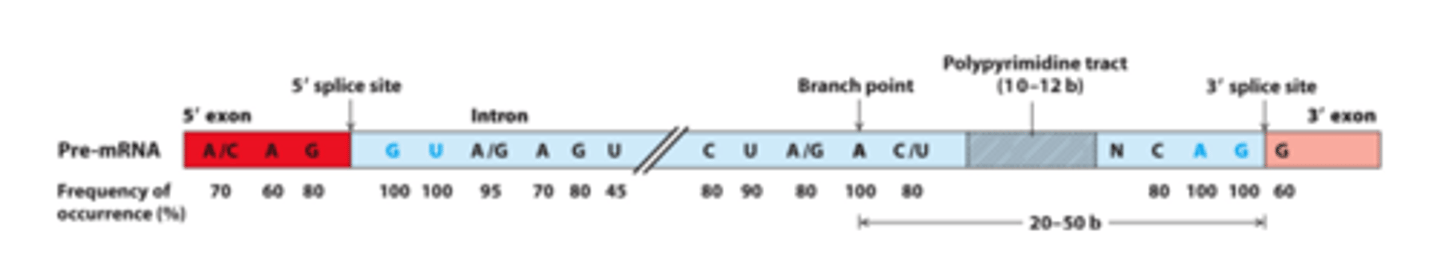
flanking bases
Intron splice site invariant bases (____________ _______ indicated - found at frequencies higher than expected for a random distribution)
branch-point
usually 20-50 bases from the 3′ splice site
GU, AG
intron splice site invariant bases: 5′ _______, 3′ ______, branch-point adenosine
polypyrimidine tract
_________________ ______ near the 3′ end of the intron - found in most introns
normal
40 bases-50 kilobases, only 30-40 nucleotides at each end of an intron are necessary for splicing to occur at ___________ rates
exon splicing
two sequential transesterification reactions [Arrows indicate where activated hydroxyl oxygens react with phosphorus atoms]
![<p>two sequential transesterification reactions [Arrows indicate where activated hydroxyl oxygens react with phosphorus atoms]</p>](https://knowt-user-attachments.s3.amazonaws.com/2a8691ba-3875-47ac-9554-f0e0258733c9.image/png)
splicing
two transesterification reactions result in the _____________ of exons in pre-mRNA
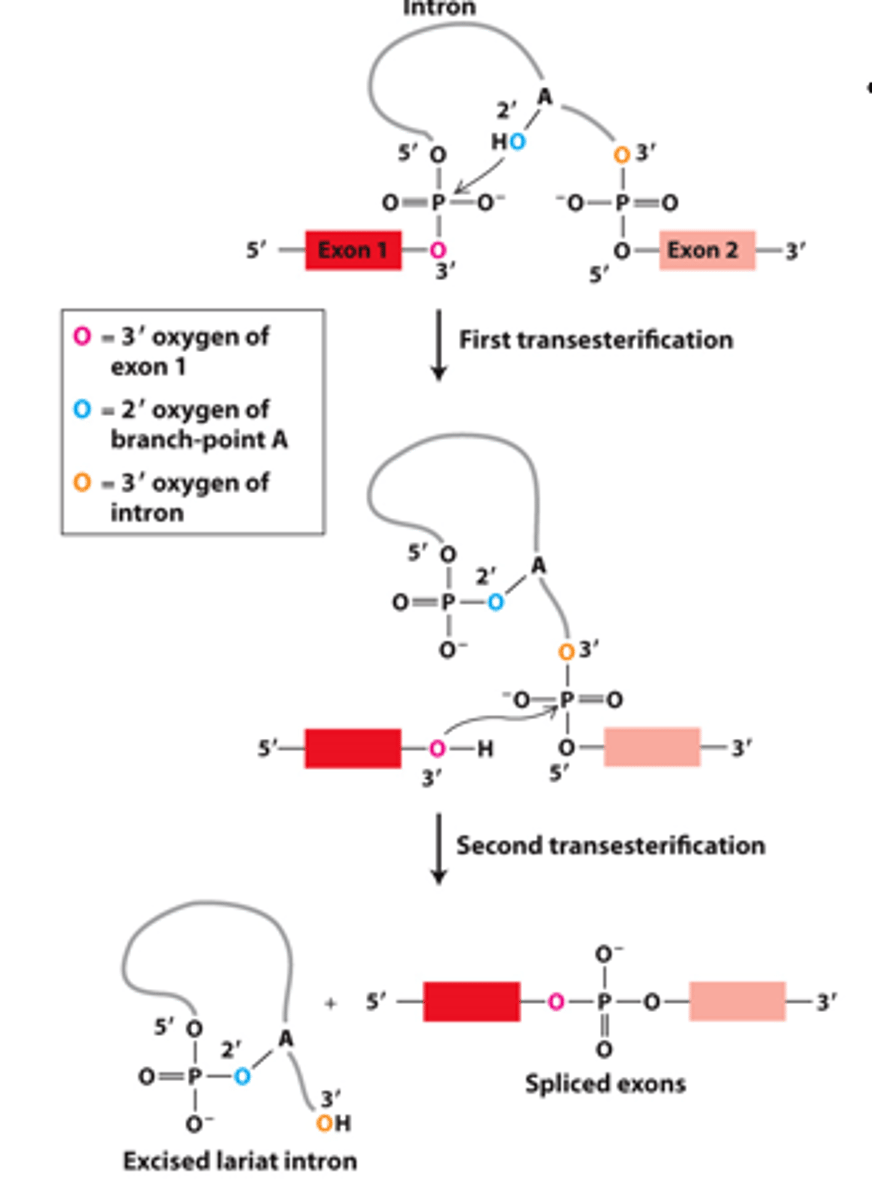
reaction 1
two transesterification reactions: ____________ _____- intron 5′ phosphorus-exon one 3′ oxygen ester bond - exchanged for an intron 5′ phosphorus ester bond with the branch-point A residue 2′ oxygen
reaction 2
two transesterification: ___________ _____ -the exon two 5′ phosphorus - intron 3′ oxygen ester bond - exchanged for an exon two 5′ phosphorus ester bond with the 3′ oxygen of exon one
joins
two transesterification: reaction 2--_______ the two exons
releases intron as a lariat structure
base pairing
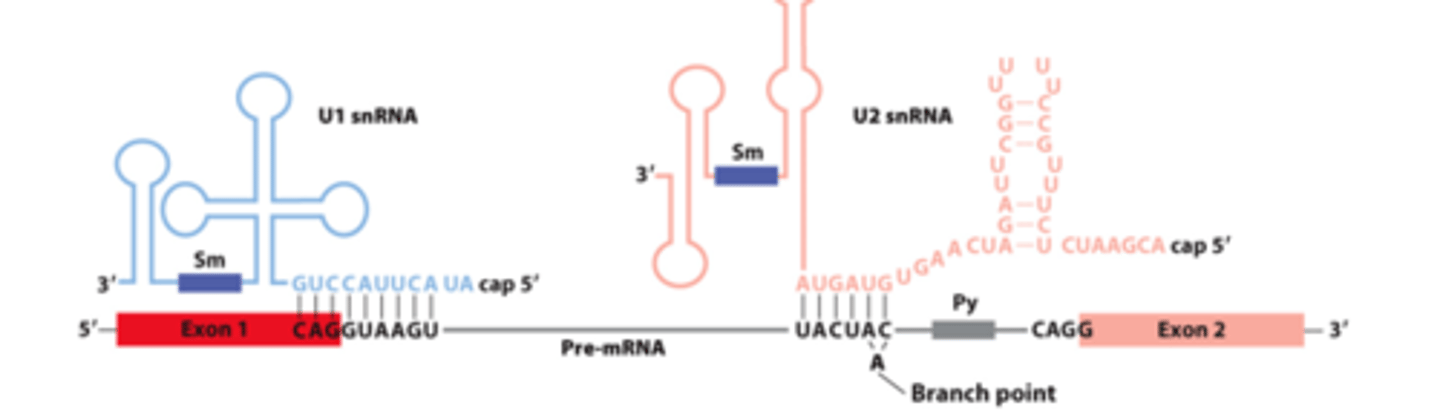
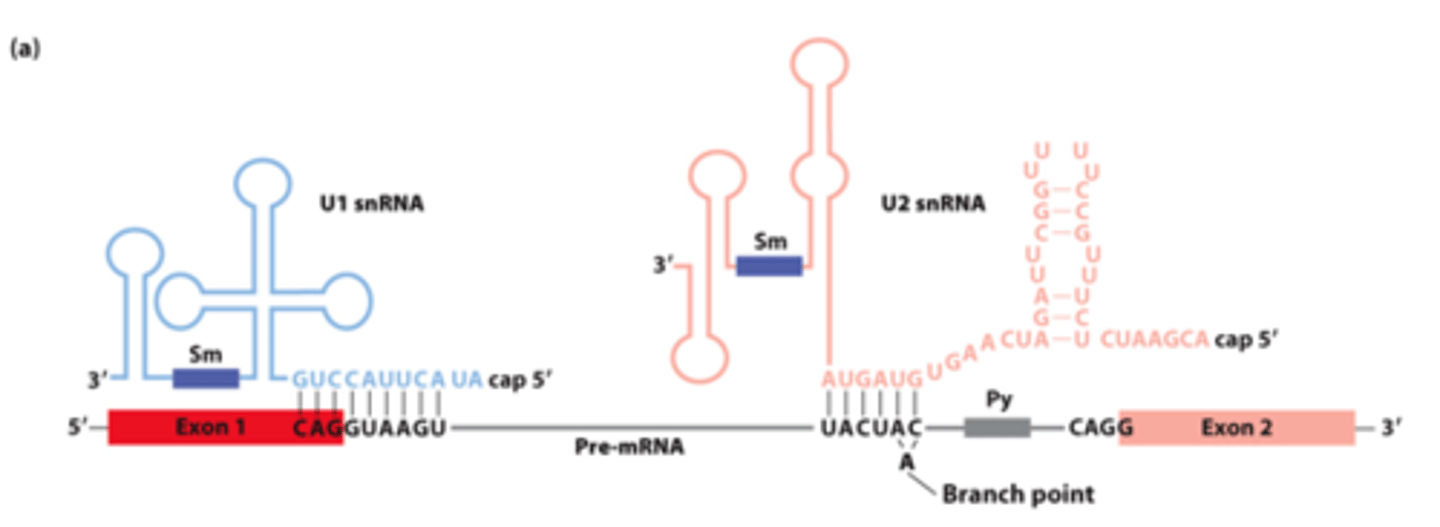
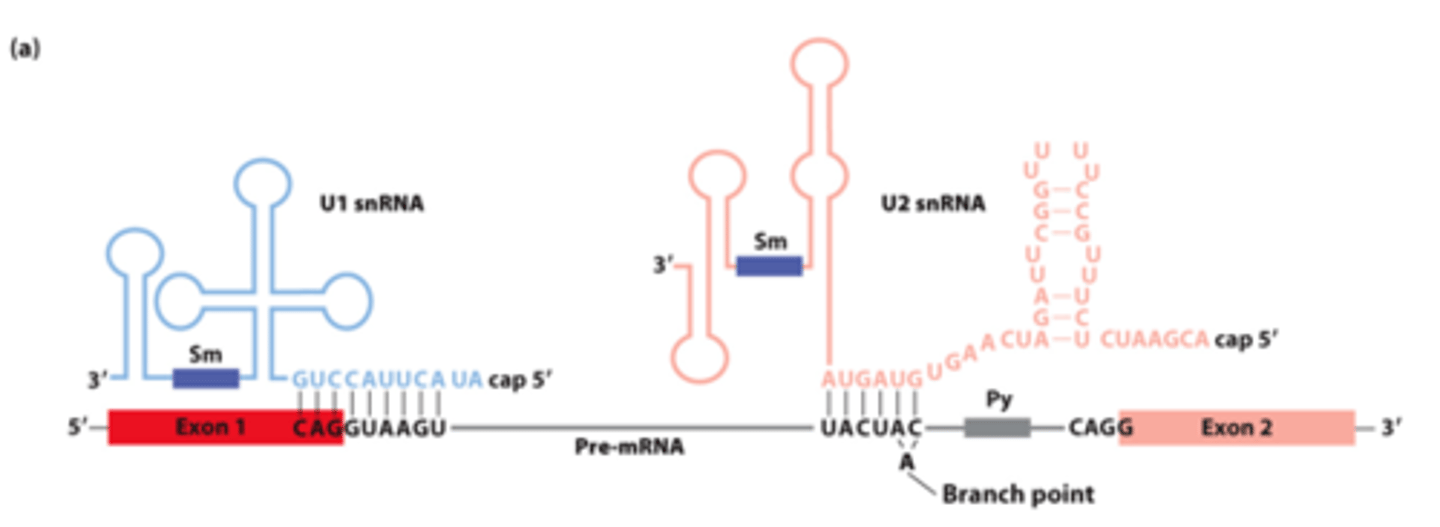
________ __________ between pre-mRNA, U1 snRNA, and U2 snRNA early in the splicing process
pre mRNA
snRNAs involved in splicing: five U-rich snRNAs - U1, U2, U4, U5, and U6 (107-210 nucleotides long) - participate in ______-_____________ splicing
base pair
snRNAs involved in splicing: ________-__________ with pre-mRNA
nuclear ribonucleoprotein particles
snRNAs involved in splicing: interact with 6-10 proteins each - form small __________ _______________ ____________ (snRNPs)
U1, U2
________ and _______ base-pairing with pre-mRNA for splicing: (purple rectangles - sequences that bind snRNP proteins recognized by anti-Sm protein antibodies)
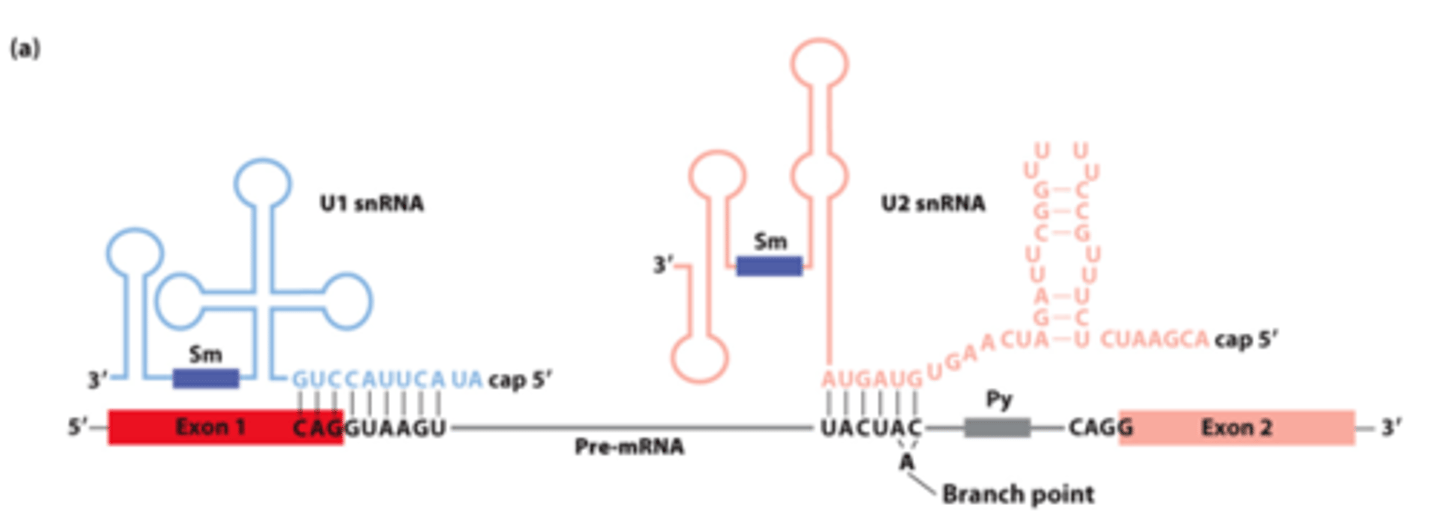
U1
U1 and U2 base-pairing with pre-mRNA for splicing: _______- base-pairs across 5' splice site exon-intron junction
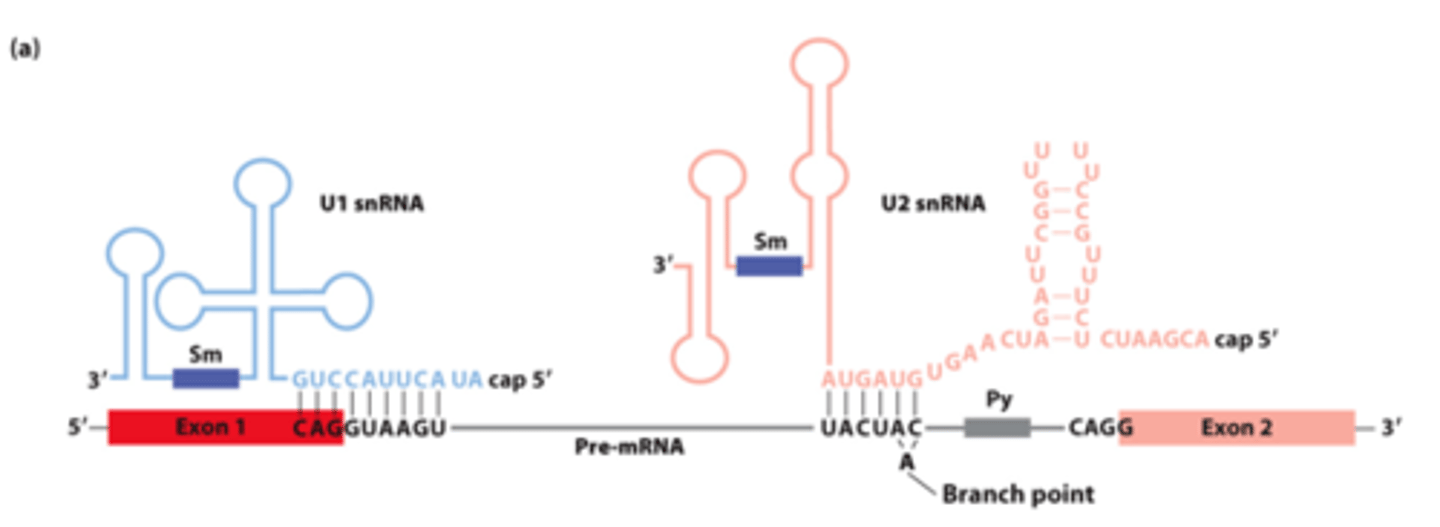
U2 snRNA
U1 and U2 base-pairing with pre-mRNA for splicing: _______ __________-- base-pairs with sequence surrounding the branch-point A, unbase-paired branch-point A bulges out to allow 2'-OH to participate in first transesterification reaction
U1 snRNA, splicing
mutations that inhibit or restore splicing: (left) – mutation (A) in a pre-mRNA splice site, interferes with base pairing to the 5′ end of ______ ________, blocks ___________
base pairing
mutations that inhibit or restore splicing: (right) – U1 snRNA with a compensating mutation (U) – restores ________ ____________, restores splicing of the mutant pre-mRNA
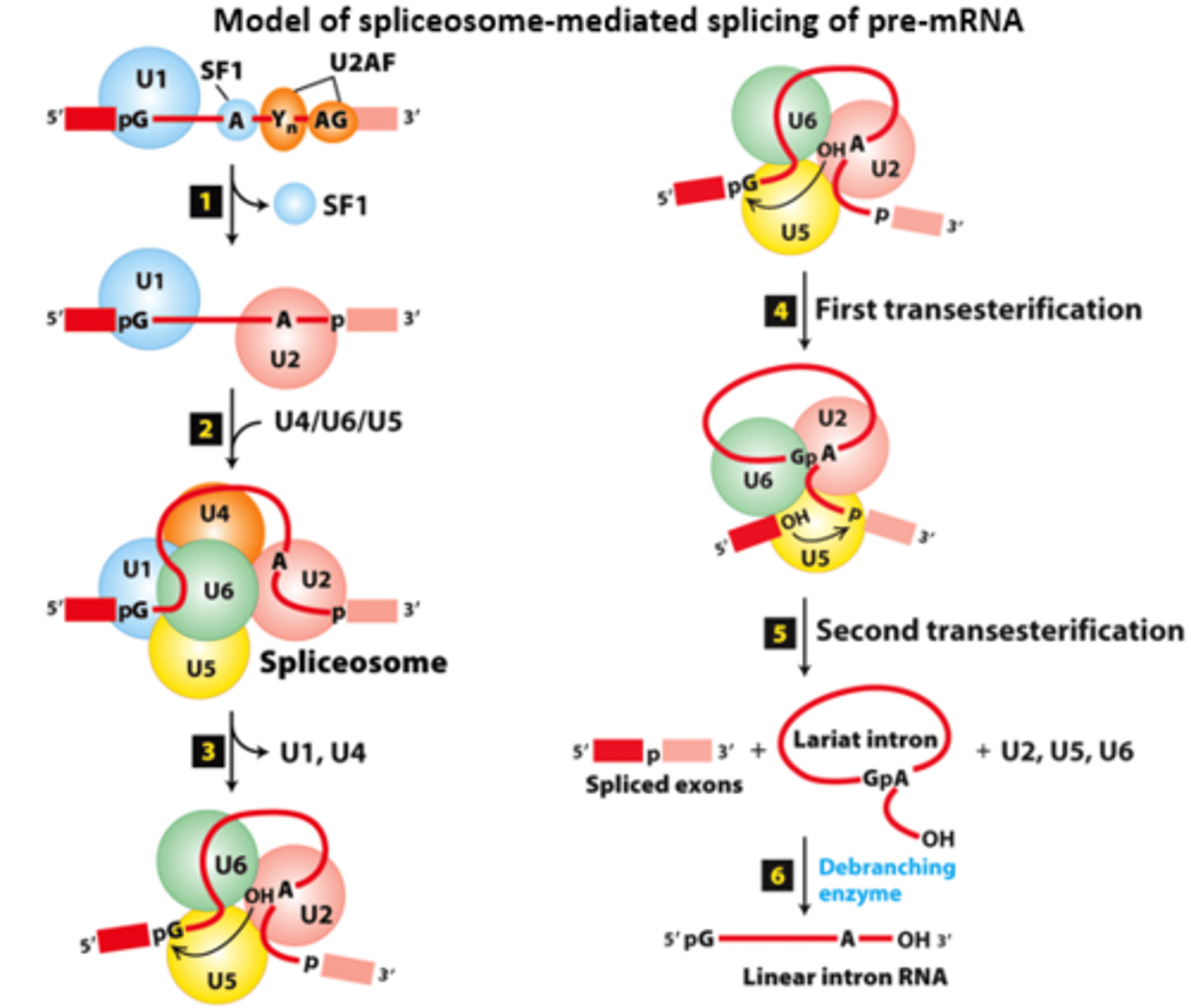
spliceosome
model of __________-mediated splicing of pre-mRNA
five U-rich snRNAs
snRNAs involved in splicing: U1, U2, U4, U5, and U6 (107–210 nucleotides long),
participate in pre-mRNA splicing
proteins
snRNAs involved in splicing: five U-rich sRNAs, base pair with pre-mRNA, interact with 6-10 __________ each
small nuclear ribonucleoprotein particles
snRNAs involved in splicing: interact with 6-10 proteins each--form _______ ____________ ______________ _______ (snRNPs)
spliceosome
five splicing snRNPs and splicing proteins assembled on a pre-mRNA
U1
spliceosome splicing mechanism, initial complex assembly:
_____ - base-pairs with the consensus 5′ splice site
splitting factor 1
spliceosome splicing mechanism, initial complex assembly: ____________ ____________ ____ (SF1) binds the branch-point A
U2 snRNP associated factor
spliceosome splicing mechanism, initial complex assembly: ______ __________ ____________ ___________ (U2AF) associates with the polypyrimidine tract and 3′ splice site
U2 snRNP
spliceosome splicing mechanism, step 1: _____ _________ - associates with the branch-point A via base-pairing interactions and displaces SF1
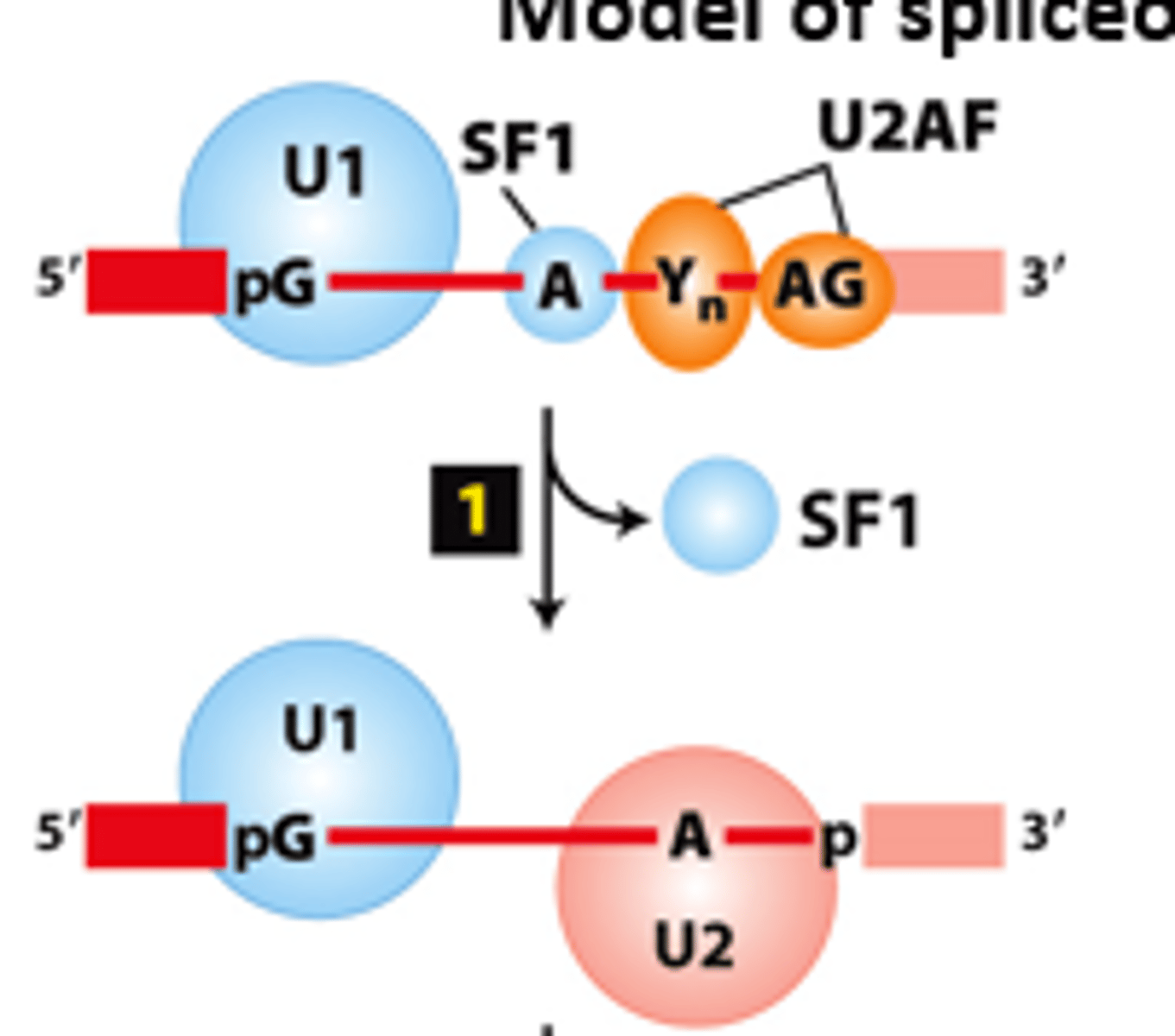
spliceosome
spliceosome splicing mechanism, step 2: U4-U5-U6 trimeric snRNP complex - binds to form the ______________
active
spliceosome splicing mechanism, step 3: snRNA base-pairing interaction rearrangements--
convert the spliceosome into a catalytically _________ conformation
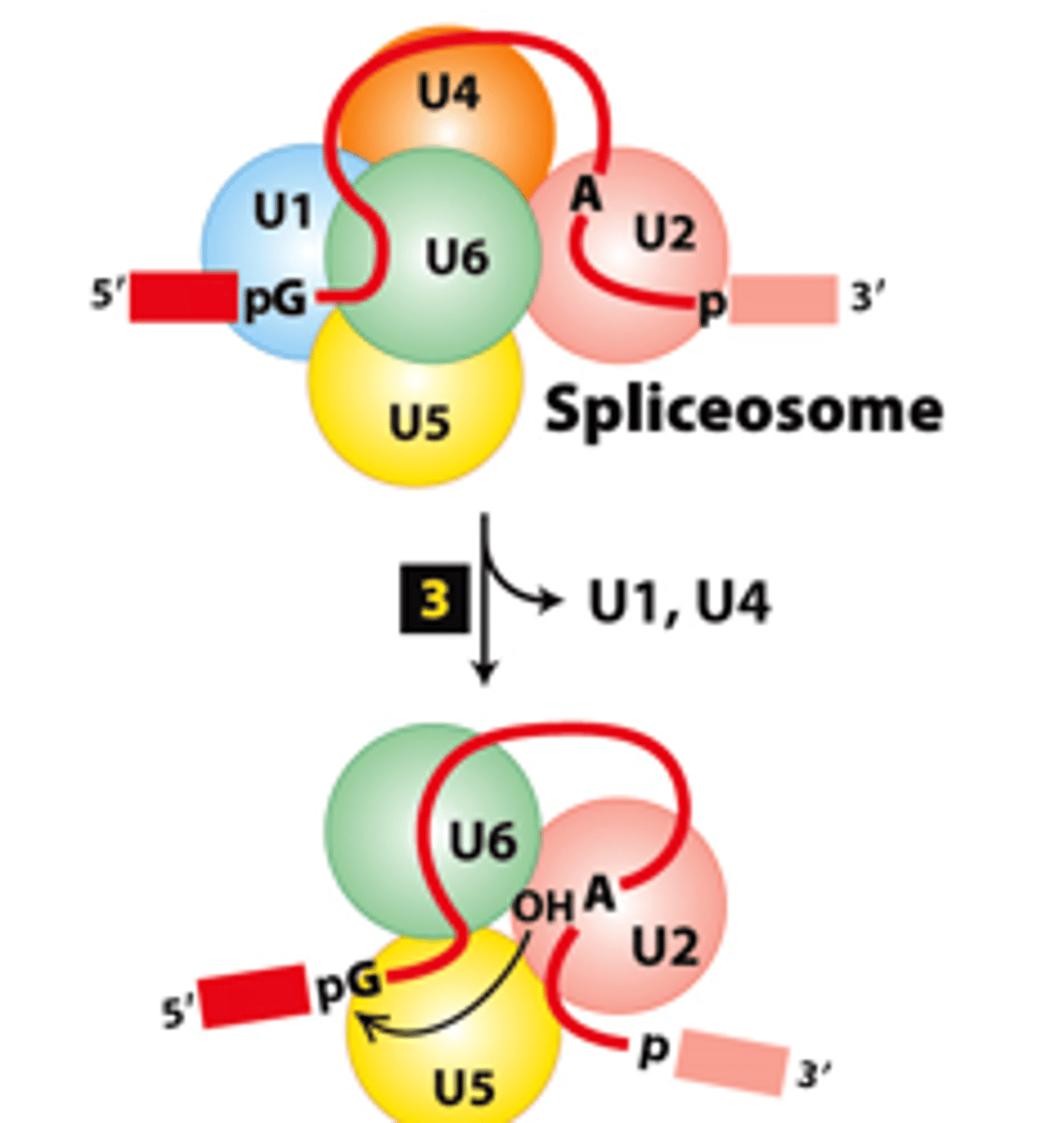
U1, U4
spliceosome splicing mechanism, step 3: snRNA base-pairing interaction rearrangements--_______ and ______ snRNPs - released
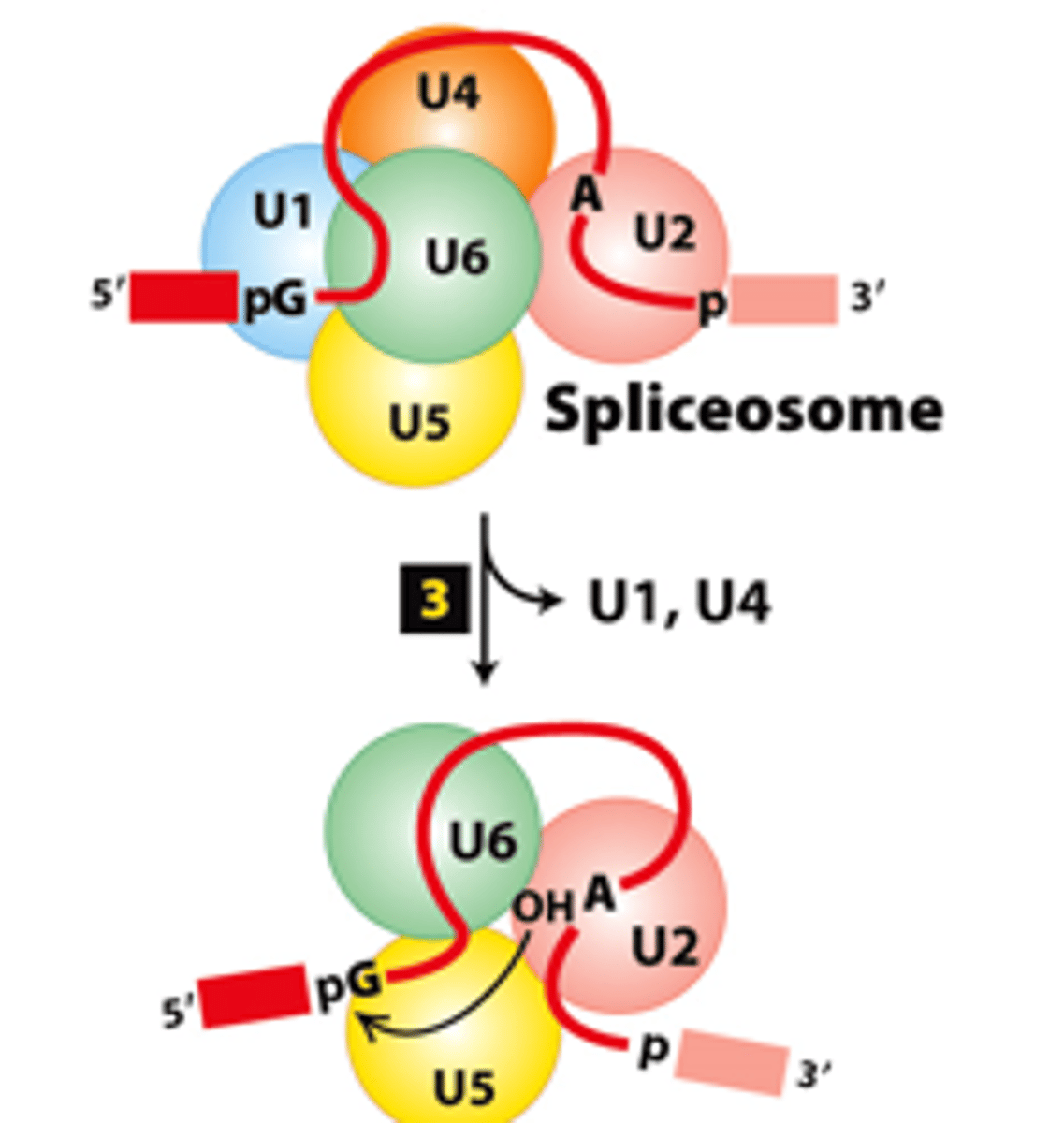
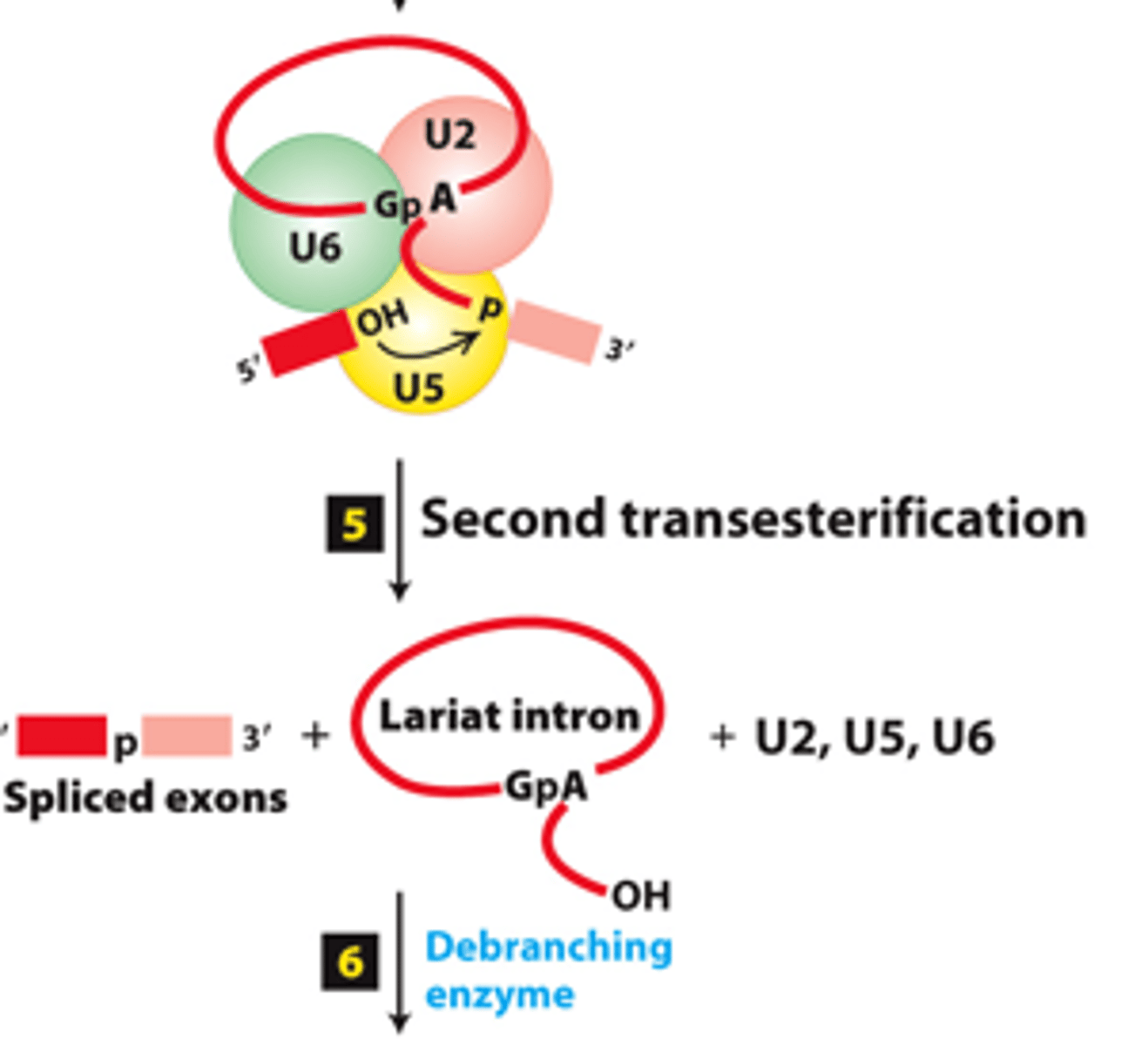
transesterification
spliceosome splicing mechanism, step 4: First _____________ reaction - U6-U2 catalytic core catalyzes formation of intermediate containing a 2′,5′-phosphodiester bond
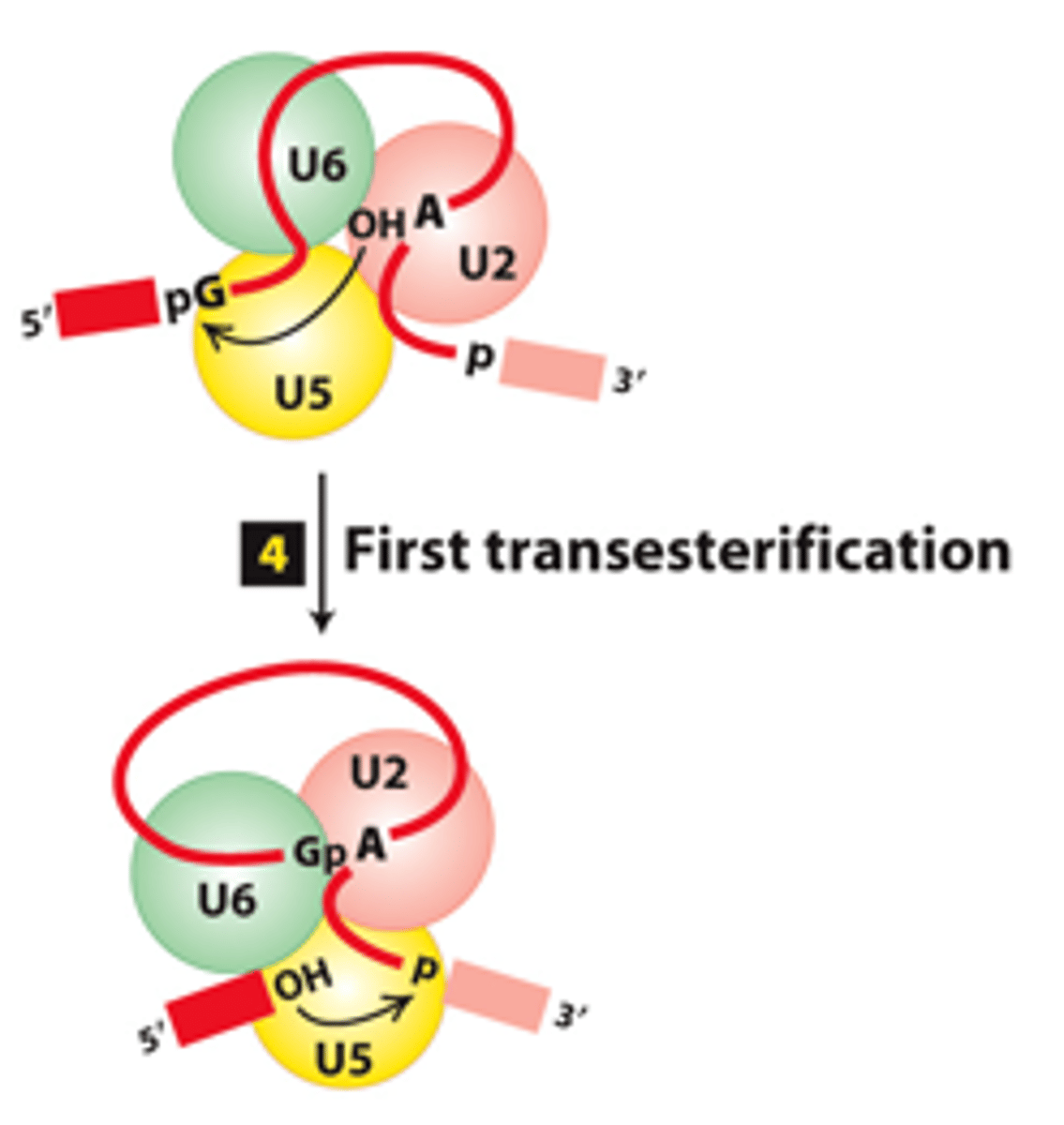
phosphodiester
spliceosome splicing mechanism, step 5: second transesterification reaction--joins the two exons by a standard 3′,5′-_____________ bond
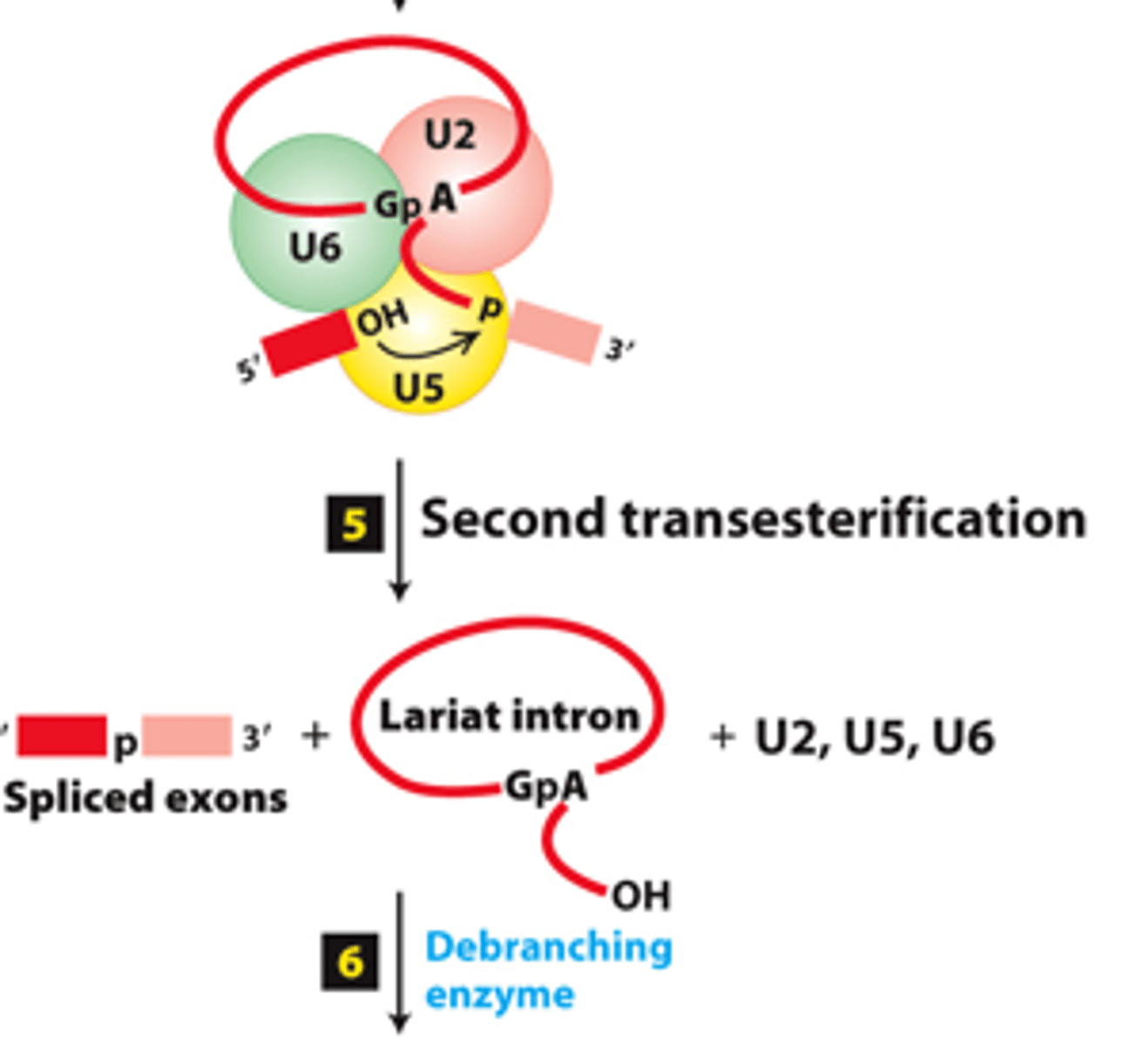
intron
spliceosome splicing mechanism, step 5: second transesterification reaction--releases the __________ as a lariat structure
releases
spliceosome splicing mechanism, step 5: second transesterification reaction--__________ remaining snRNPs
linear RNA
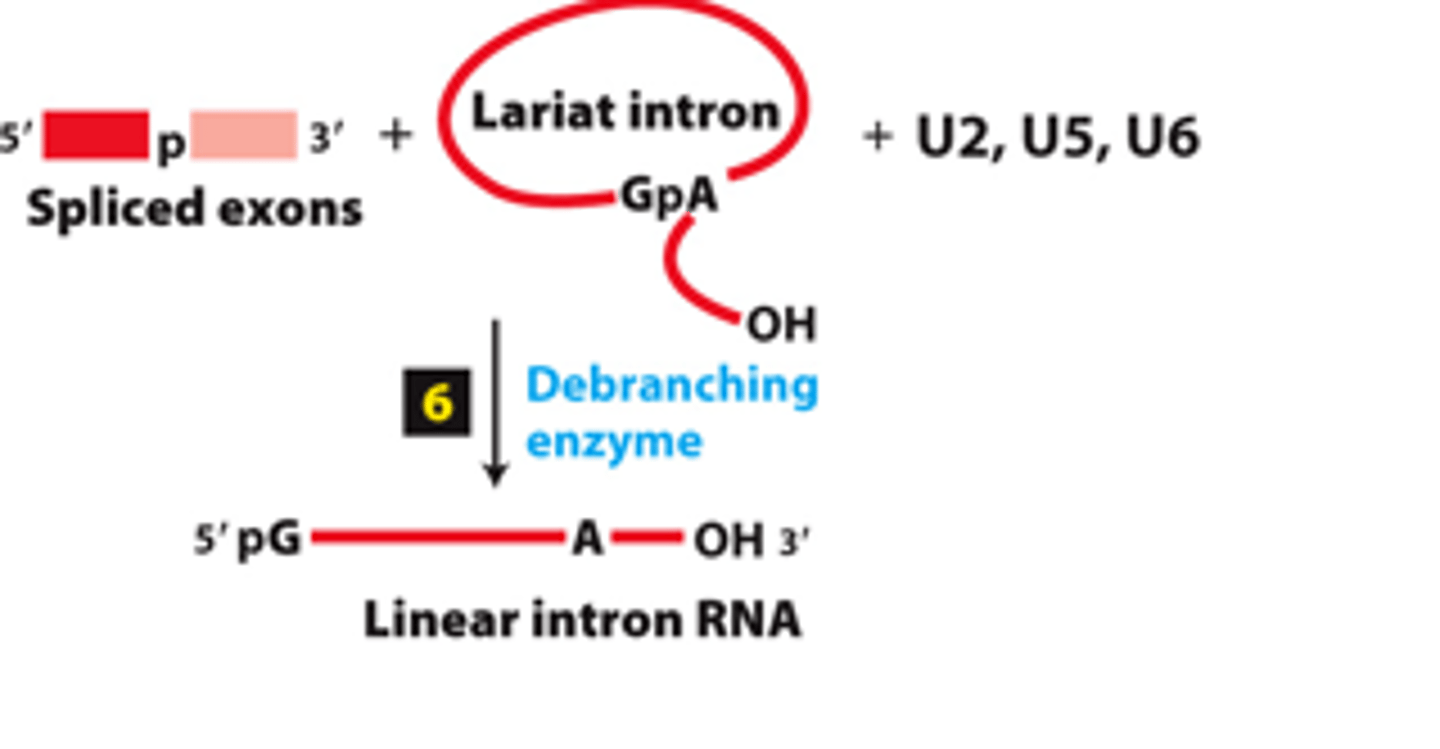
spliceosome splicing mechanism, step 6: debranching enzyme coverts excised lariat intron into a __________ _______for degradation
cooperative binding
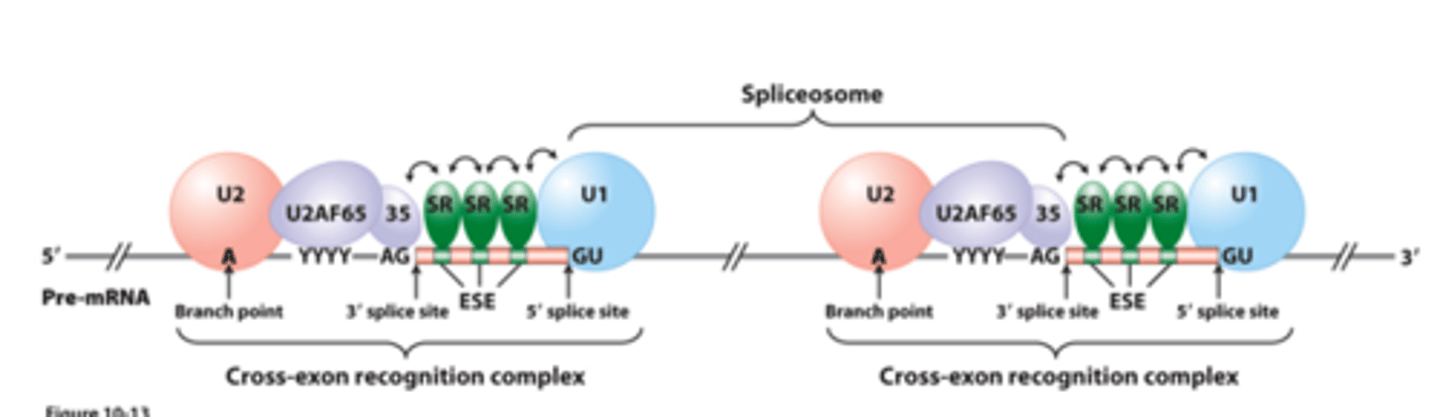
exon recognition through _____________ ___________ of SR proteins and splicing factors to pre-mRNA
sr proteins
named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are "S" and "R" respectively
sr proteins
interact with exonic enhancer sequences (ESEs) within exons
sr proteins
contribute to exon definition in long pre-mRNAs
exons
pre-mRNAs humans: avg ~150 bases
introns
pre-mRNAs humans: avg ~3500 bases - longest exceed 500 kb
degenerate
pre-mRNAs humans: _____________ 5′ and 3′ splice site and branch point sequences - multiple copies likely to occur randomly in long introns
spliced together
pre-mRNAs humans: additional sequence information is required to define the exons that should be _________ ______________ in higher organism pre-mRNAs with long introns
arginine, serine
SR proteins: contain several RS protein-protein interaction domains rich in ___________ (R) and ___________ (S) residues
exonic enhancer sequences
SR proteins:
interact with _________ ___________ ____________ (ESEs) within exons
protein protein
SR proteins: mediate cooperative binding of U1 snRNP to a true 5′ splice site and U2 snRNP to a branch point through a network of ___________-____________ interactions that span an exon
GU, AG
5' _______ and 3' _________ splice sites recognized by splicing factors on the basis of their proximity to exons
interact
SR proteins bound to ESEs: ________ with each other
downstream
SR proteins bound to ESEs: promote cooperative binding of--U1 snRNP to the 5′ splice site of the ______________ intron
upstream
SR proteins bound to ESEs: promote cooperative binding of--U2 snRNP binding to the branch point of the upstream intron, the 65- and 35-kDa subunits of U2AF to the polypyrimidine tract and AG 3′ splice site of the ____________ intron, and other splicing factors
protein cross exon
resulting RNA-________ ____________-_________ recognition complex - spans an exon and activates the correct splice sites for RNA splicing
spinal muscular atrophy
one of the most common genetic causes of childhood mortality
identical
SMN1 and SMN2 genes--encode ___________ proteins
silent
SMN2 ____________ mutation interferes with SR protein binding