Cell BIO Exam 2
1/342
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
343 Terms
What is Gene expression?
The process of turning DNA into protein through multiple steps.
What is transcription?
The process of transcribing a piece of DNA into RNA.
What is pre-messenger RNA?
RNA synthesized in eukaryotes that is immature and needs processing.
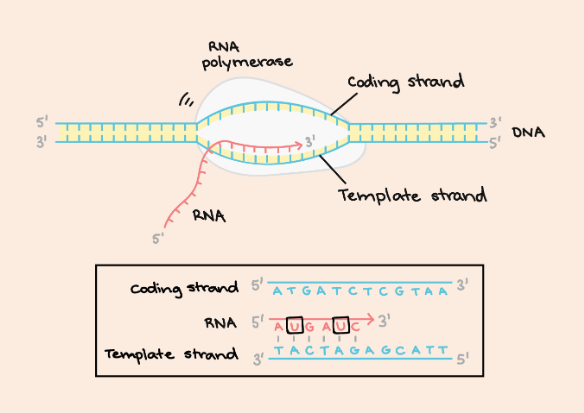
What is the coding strand?
The non-template strand; the strand that is identical to the newly synthesized RNA (except U & T)
What is a promoter?
Specific sequence on the template DNA that RNA polymerase locates to start transcription.
What does a sigma factor do?
Helps RNA polymerase recognize the promoter on the gene.
What are terminator sequences?
Sequences at the end of the gene that tell RNA polymerase to stop.
What does TFIID do?
Binds to the double-stranded DNA in eukaryotic transcription.
What does the TATA-binding protein do?
Recognizes the TATA box promoter sequence.
What are introns?
Parts of the RNA that are removed during RNA splicing.
What does splicing do?
Joins exons together by removing introns.
What does alternate splicing do?
Produces different messenger RNAs from the same gene by selectively removing different exons.
What must happen before transcription to occur?
The DNA must unwind and the promoter region must be accessible for RNA polymerase (heterochromatin → euchromatin)
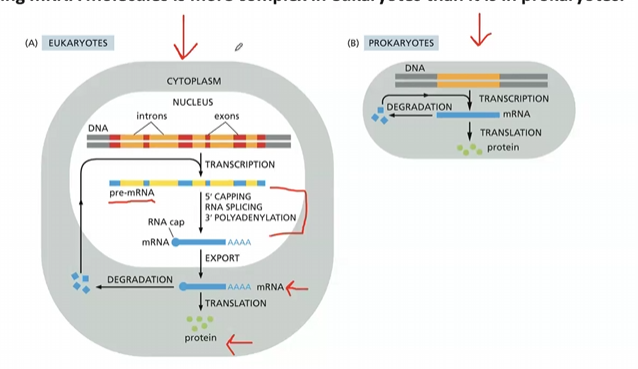
Is producing mRNA molecules more complex in eukaryotes or prokaryotes? Why?
Producing mRNA molecules is more complex in eukaryotes because they undergo extensive post-transcriptional modifications, including capping, polyadenylation, and splicing, whereas prokaryotes have simpler transcription processes that do not involve these modifications.
How do RNA and DNA differ?
RNA contains ribose sugar and uracil, while DNA contains deoxyribose sugar and thymine.
RNAs that code for proteins
mRNAs
RNAs that form the core of the ribosome’s structure and catalyze protein synthesis
rRNAs
RNAs that regulate gene expression
mRNAs
RNAs that serve as adaptors between mRNA and amino acids during protein synthesis
tRNAs
what are some functions of other noncoding RNAs?
RNA splicing, gene regulation, telomere maintenance, and many other processes
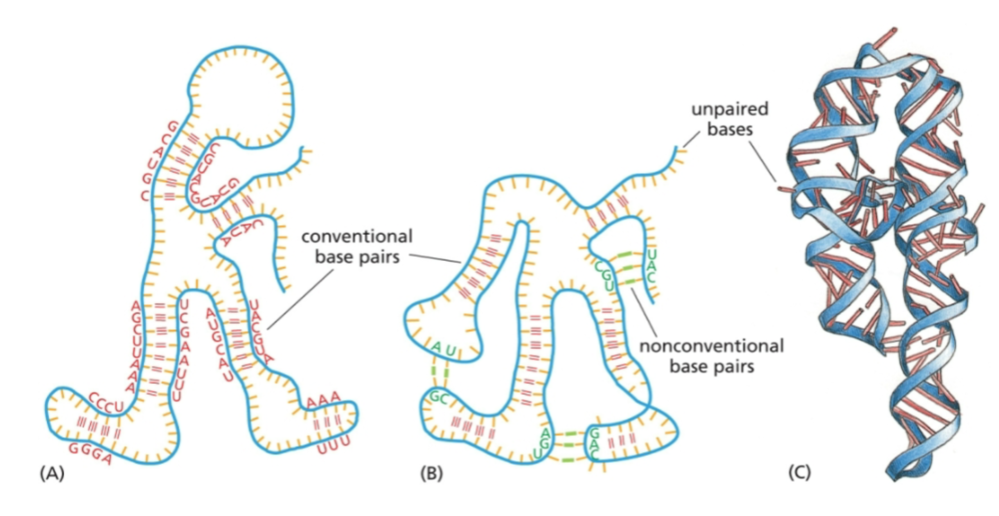
RNA molecules can fold into specific structures that are held together by…
hydrogen bonds between nucleotides
Which RNA polymerase transcribes most rRNA genes?
RNA polymerase I
Which RNA polymerase transcribes all protein-coding genes, miRNA genes, plus genes for other noncoding RNAs
RNA polymerase II
Which RNA polymerase transcribes tRNA genes, 5S rRNA gene, and genes for many other small RNAs?
RNA polymerase III
How many RNA polymerases do prokaryotes have?
only one
all RNA polymerases are made of what subunits?
two large subunits and multiple small subunits.
What step of mRNA production in eukaryotes is different than prokaryotic mRNA production?
eukaryotic mRNA production involves processing events such as capping, polyadenylation, and splicing, which do not occur in prokaryotes. 0
strand of DNA that is complementary to the newly synthesized RNA during transcription
template strand
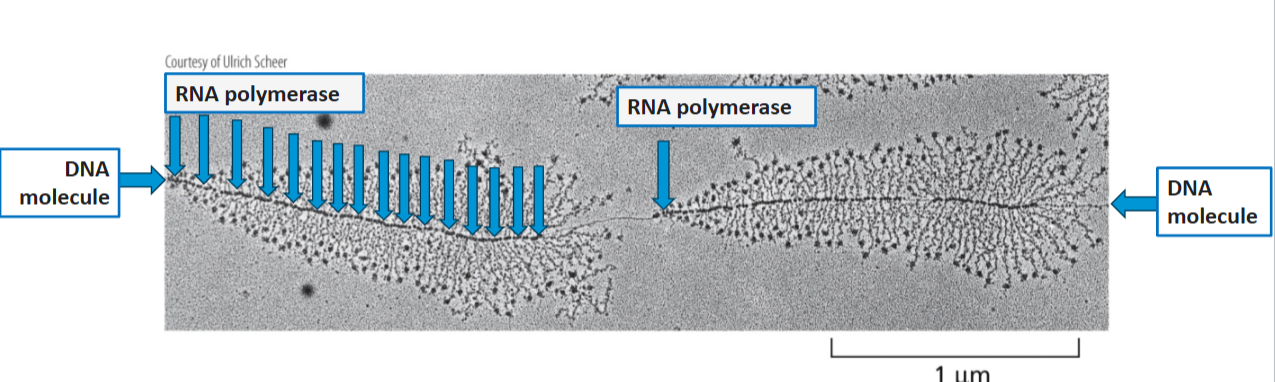
T/F: many molecules of RNA polymerase can simultaneously transcribe the same gene
true
transcription starts on the ____ end of the DNA template
3’
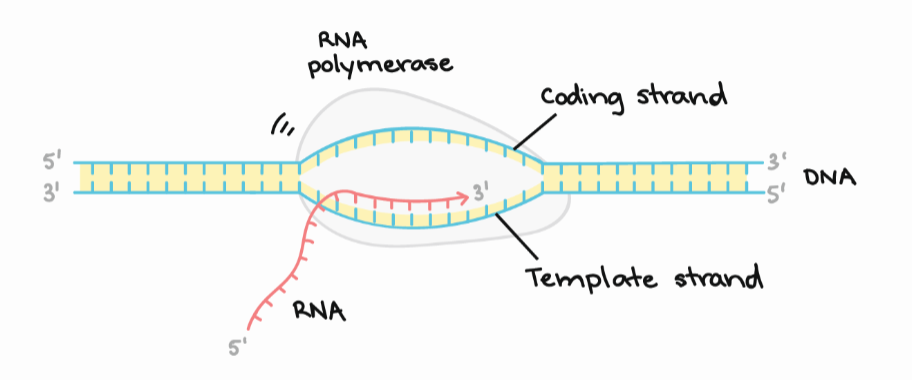
How does RNA polymerase locate where to start transcription?
by recognizing specific DNA sequences called promoters, which are located upstream of the gene to be transcribed.
subunit of bacterial RNA polymerase that recognizes the promoter of a gene
sigma factor
How does RNA polymerase know when to stop transcription?
the terminator (specific nucleotide sequence)
in what kinds of organisms are promoters found?
in both eukaryotic and prokaryotic
What is the first base that gets transcribed during transcription (AKA the first base of the RNA synthesized)?
the +1 site
In bacterial transcription, do the regions transcribed into RNA include the promoter and/or terminator?
contains only the terminator (although eukaryotic transcription can contain promoters)
where are promoters found in eukaryotes and not in prokaryotes?
downstream of the transcription start site (aka within the transcription region)
where are eukaryotic mRNAs processed?
in the nucleus
where are ribosomal RNAs synthesized and combined with proteins to form ribosomes?
in the nucleolus
Where do mRNA molecules go after being synthesized and how?
to the cytosol via pores in the nuclear envelope
three steps of transcriptional and post-transcriptional modifications
capping at the 5’ end
addition of poly-A tail at the 3’ end
RNA splicing
noncoding sequences that interrupt protein-coding genes in eukaryotes
introns
How are introns removed from pre-mRNAs?
RNA splicing
There are five _____ and about 200 additional proteins, called a _____, required for splicing
snRNPs, spliceosome
the general transcription factors that assemble at the promoter are the same/different for all (eukaryotic) genes
the same
the transcription regulators and the locations of their binding sites are the same/different for different genes?
different
protein that attaches to RNA polymerase and assembles all of the transcription factors and regulators
mediator
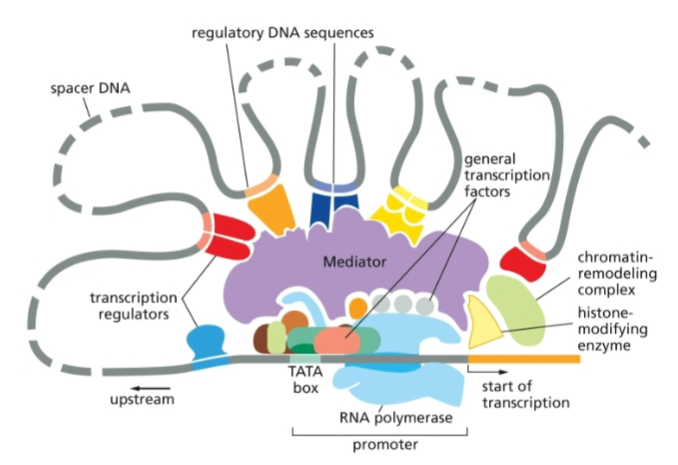
transcription factor that recognizes and binds to the TATA box, where it then recruits other transcription factors
TFIID aka TATA-binding protein (TBP)
DNA sequence of eukaryotic promoters 30 bp upstream of the start site
TATA box
What is the purpose of capping the 5’end of mRNA?
to protect it from degradation, help it exit the nucleus, and allow ribosomes to recognize and bind to it for translation.
What is the purpose of adding the poly-A tail to the 3’ end?
helps protect it from degradation, aids in its export from the nucleus, and improves translation efficiency.
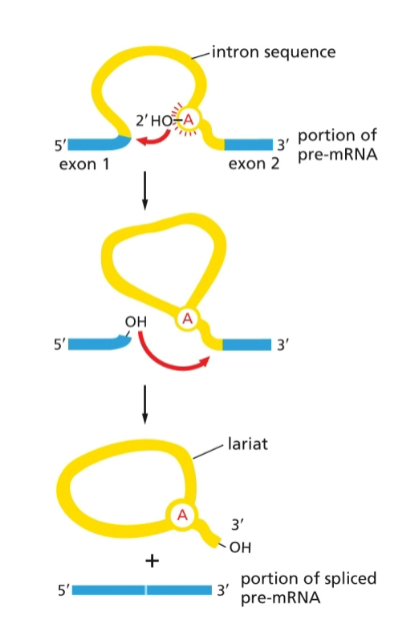
describe the process of RNA splicing in 6 steps
snRNPs form a spliceosome which binds near the 5’ splice site
spliceosome cuts at the 5’ splice site
free 5’ end bonds to the branch point A, forming a lariat
spliceosome cuts at the 3’ splice site
the free 3’-OH end of the exon sequence reacts with the start of the next exon sequence
intron lariat is released and eventually broken down
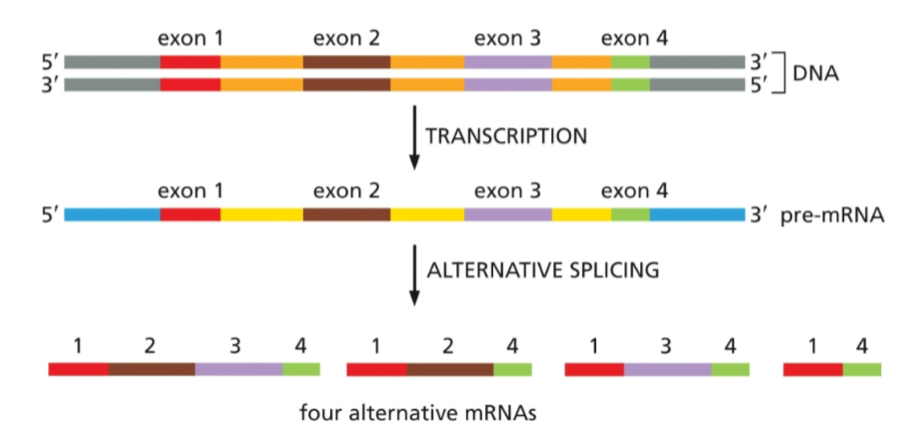
process some pre-mRNAs undergo to produce different mRNAs and proteins from the same gene
alternative splicing
what does alternative splicing allow that normal RNA splicing does not?
allows exons to be skipped or included depending on how it is spliced; although the DNA sequence can not be rearranged - ultimately allows different proteins to be made from the same gene
protein markers placed on exon-exon boundaries after successful splicing
exon junction complexes
what associates with mature eukaryotic mRNA to guide it through the nuclear pore out of the nucleus?
a nuclear transport receptor
what kinds of cells contain all the genetic instructions needed to direct the formation of a complete organism?
differentiated cells
when in the process of making DNA → RNA → protein is gene expression regulated?
at various steps throughout the process
functioning unit of bacterial DNA containing a cluster of genes under the control of a single promoter
operon
what allows bacterial cells to respond to changes in their environment by controlling operons?
transcription switches
how many genes are included in the operon needed to synthesize the amino acid tryptophan form simpler molecular building blocks
five
single regulatory DNA sequence that controls the tryptophan operon?
Trp operator (inside of the promoter region)
How does the tryptophan operon work at low concentration of tryptophan inside the cell?
RNA polymerase binds to the promoter
transcribes the five genes of the tryptophan operon, allowing production of tryptophan
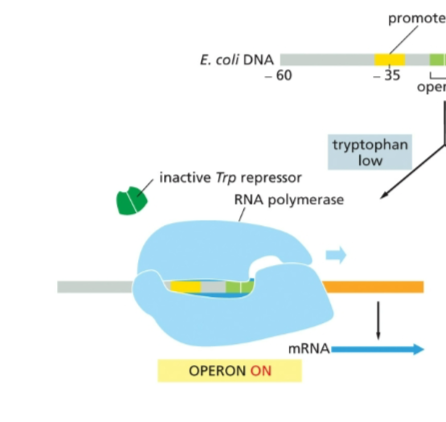
How does the tryptophan operon work at high concentration of tryptophan inside the cell?
Trp repressor binds to the operator, blocking the binding of RNA polymerase to the promoter
this prevents transcription of the operon
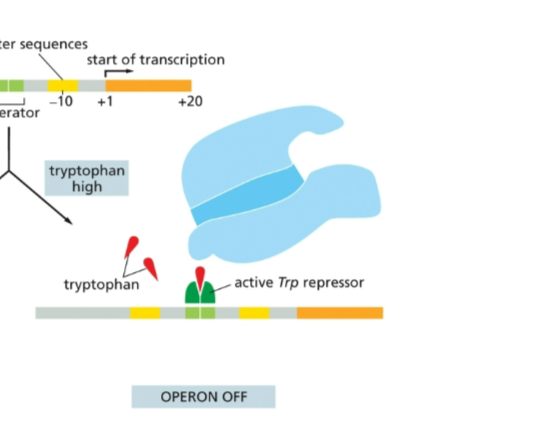
How does the Lac operon work when lactose is absent?
the Lac repressor binds to the Lac operator and shuts off expression of the operon
operon required for the transport and metabolism of lactose in E. coli and other enteric bacteria?
Lac operon
when is the Lac operon transcribed?
glucose is absent (allowing the CAP (cyclic AMP) activator to bind
lactose is present (releasing the Lac repressor)
what does the Lac operon mRNA encode for that breaks down lactose into galactose and glucose?
the enzyme β-galactosidase
eukaryotic protein that is bound to a distant enhancer that attracts RNA polymerase and the general TFs to the promoter
activator proteins
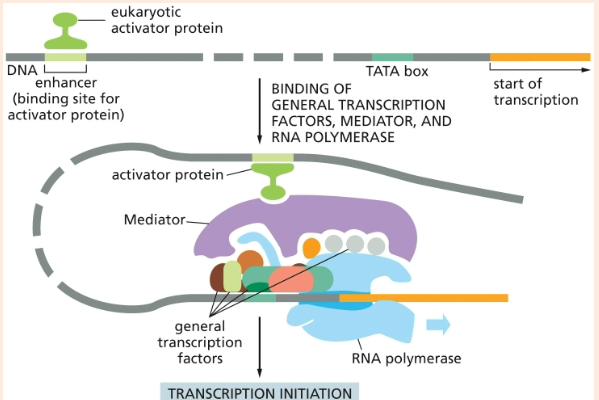
proteins that help initiate transcription by recruiting chromatin-modifying proteins
eukaryotic transcriptional activators
chromatin-modifying protein that adds acetyl groups to histones to stimulate transcription initiation
histone acetyltransferases
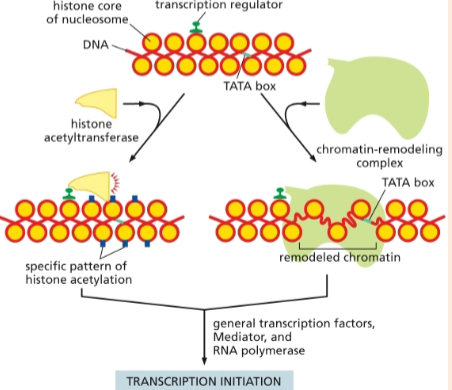
chromatin-modifying proteins that make the DNA more accessible to other proteins in the cell; required for transcription initiation
chromatin-remodeling complexes
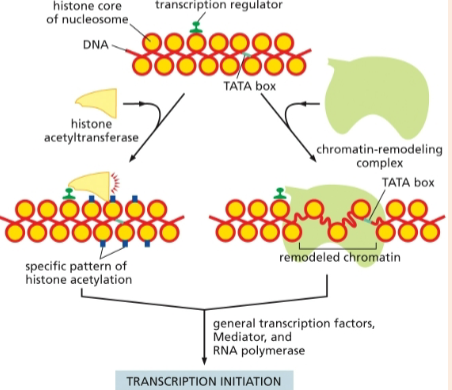
location of gene transcription and translation in prokaryotes and eukaryotes
prokaryotes: coupled; no nucleoid envelope barrier because prokaryotic cell structure
eukaryotes: nuclear transcription and cytoplasmic translation
describe gene clustering of prokaryotes and eukaryotes
prokaryotes: operons where genes with similar functions are grouped together
eukaryotes: operons generally not found; each gene has its own promoter element and enhancer elements
default state of transcription in prokaryotes vs eukaryotes
prokaryotes: on
eukaryotes: off
difference in DNA structure between prokaryotes and eukaryotes?
eukaryotic DNA is associated with histones in nucleosomes, while prokaryotic DNA has some other associated proteins, but not histones
the area of the chromosome which is rich in genes that actively participate in the transcription process
euchromatin
the area of the chromosome which is darkly stained with a DNA-specific stain and is in comparatively condensed form
heterochromatin
A regulatory DNA sequence that, when bound by specific proteins called transcription factors, enhances the transcription of an associated gene
enhancer
How did Nirenberg and Matthaei discover the code for phenylaline?
they produced synthetic mRNAs made entirely of uracil (poly U). Poly U was then fed to a translation system containing bacterial ribosomes, and found that it only synthesized a polypeptide containing only phenylalanine (meaning the code for Phe must be UUU)
how did researches make a cell-free system for protein synthesis
centrifuged broken E.coli
added radioactive amino acids to trigger the production of radiolabeled polypeptides
after centrifuging again, the labeled polypeptides could be detected by measuring radioactivity
stop codons
UAA, UAG, UGA
what part of a tRNA molecule binds to a complementary mRNA codon
the anticodon
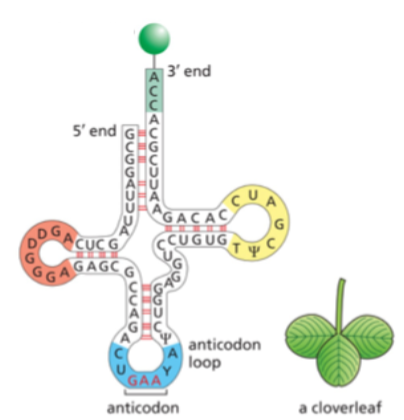
what end of the tRNA does the amino acid matching the anticodon attach to?
the 3’ end
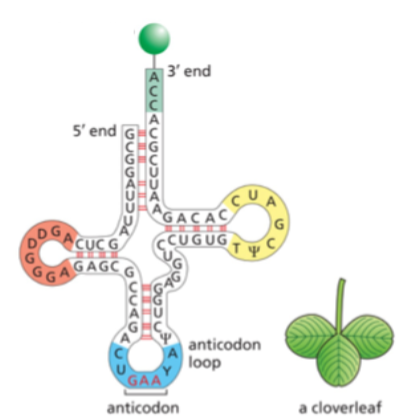
2 unusual bases found in tRNAs that are produced by chemical modification after the tRNA has been synthesized
ψ (for pseudouridine) and D (for dihydrouridine) are
derived from uracil
in principle, in how many different possible reading frames can an mRNA molecule be translated?
3
Large macromolecular complexes, composed of RNAs and proteins, that translate messenger RNAs into a polypeptide chains
ribosomes
what do the small (40s) and large (60s)ribosomal subunits do?
small subunit matches the tRNAs to the codons of the mRNA
large subunit catalyzes the the formation of the peptide bonds that covalently link the amino acids together in the polypeptide chain
four repeating steps of translation
charged tRNA carrying aa binds to vacant A site
carboxyl end of P site aa is uncoupled from tRNA at the and joined by a peptide bond to the free amino group of the amino acid linked to the tRNA at the A site
a shift of the large subunit relative to the small subunit moves the two bound tRNAs into the E and P sites of the large subunit
small subunit moves three nucleotides along the mRNA molecule, bringing it back to its original position relative to the large subunit; ejects the spent tRNA
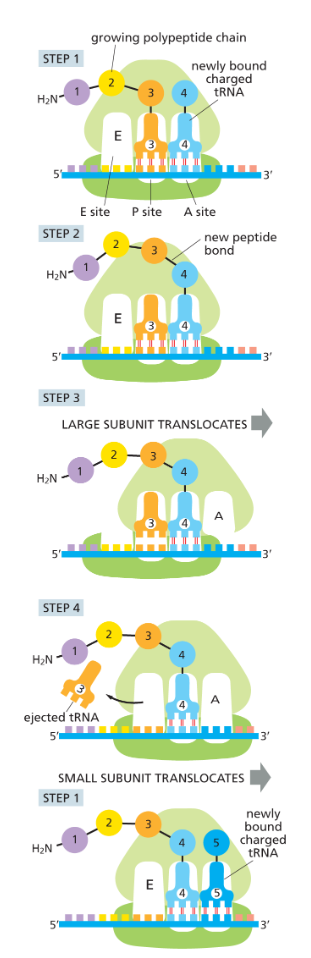
what are the three binding sites in each ribosome?
A site - aminoacyl-tRNA
P site - peptidyl-tRNA
E site - exit
two molecules required for protein synthesis initiation in eukaryotes
translation initiation factors (eIF1-3)
a special initiator tRNA (methionyl-tRNA - Met-tRNAi)
can a single prokaryotic mRNA molecule encode several different proteins?
yes
a series of ribosomes that can simultaneously translate the same mRNA molecule in both prokaryotic and eukaryotic mRNAs
polyribosomes (polysomes)
protein in eukaryotic polysomes that bind to the 3’ poly-A tail which loops the mRNA into a circular shape; promoting efficient re-initiation of translation, allowing continuous protein synthesis, and contributing to stability of the mRNA
poly-A binding protein (PABPI)
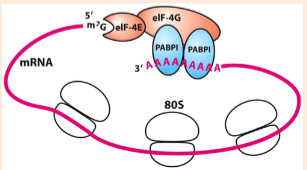
Even after an mRNA and its corresponding protein have been produced, their concentrations can be regulated by _____
degradation
how do cells use degradation to regulate the amount of protein in a cell?
proteins marked by a polyubiquitin chain are degraded by a proteasome
proteasomes are lined with proteases that chop the protein to pieces
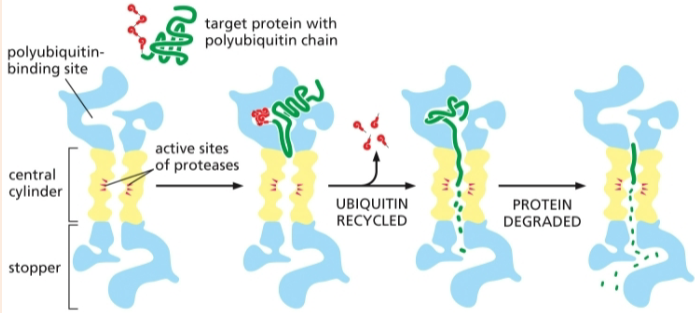
what are some things a protein may need to do to become fully functional after translation?
fold correctly into its 3D conformation
bind required cofactors and other proteins
may need covalent modifications (phosphorylation, glycosylation, etc.)
antibiotic that blocks binding of aminoacyl-tRNA to A site of ribosome
tetracycline