11. Bacterial transcription
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
24 Terms
Gene expression can be regulated at the level of __ or __ or both
Transcription
Translation
The mRNA is called the ‘sense strand’ so when it is replicated, the non template and template strand are called what?
Non-template = sense strand cause its the same as the mRNA one but with T instead of U. (You can translate the info from it as it is the same as the mRNA)
Template strand = antisense strand because it is coded for using the mRNA so isn’t the same as the mRNA.
Why is uracil not found in DNA?
Cytosine can undergo spontaneous deamination to produce uracil and DNA replication after this would replace a C-G base pair with a U-A base pair introducing mutations. This is why in DNA any U is removed by uracil-DNA glycosylase (removes the sugar), generating an a basic site which is removed and repaired by DNA polymerases.
List the three major classes of bacterial RNA:
MRNA - encodes proteins
rRNA - constituents of ribosomes (role in protein synthesis)
tRNA - adaptors between mRNA and amino acids (role in protein synthesis).
(In E.coli only one polymerase makes all of these)
What does the sense strand look like in bacterial transcription?
There is a promoter (controlled by an operator) on the 5’ end of the sense strand. (where pol binds)
There is a transcribed protein coding sequence (often multiple genes that is polycistronic as part of an operon.
There is a 3’ terminator that signals the stop point for transcription.
Why do transcription and translation happen at the same time in bacteria?
There is no nucleus.
What is bacterial RNA polymerase made up of?
alpha, beta, omega and sigma subunits. Ratio 2:1:1:1
Sigma provide the specificity.
During transcription, there is a non specific sliding RNA polymerase that can’t recognise the promotor region. How does it recongnise it?
It has a sigma factor that binds to the core polymerase and recognises the promotor and stops when it does.
What is DNA footprinting for?
It is used to identify promoters as when they are in the holoenzyme RNA polymerase, they are unreachable to DNAase.
How does DNA footprinting work?
Radioactively label the 3’ end of DNA with a phosphate
Add some DNAse and it will nick and break off different length fragments of DNA.
Electrophorese the nested ladder of phosphate labelled fragments.
How do you use this to work out where a promoter is?
Add the holoenzyme RNA pol that binds to the promotor.
Add the DNAse to cut the DNA
The pol means that the DNA is inaccessible to the DNAse so you don’t get cut fragments where the promoter is.
This will create a gap in the electrophoresis.
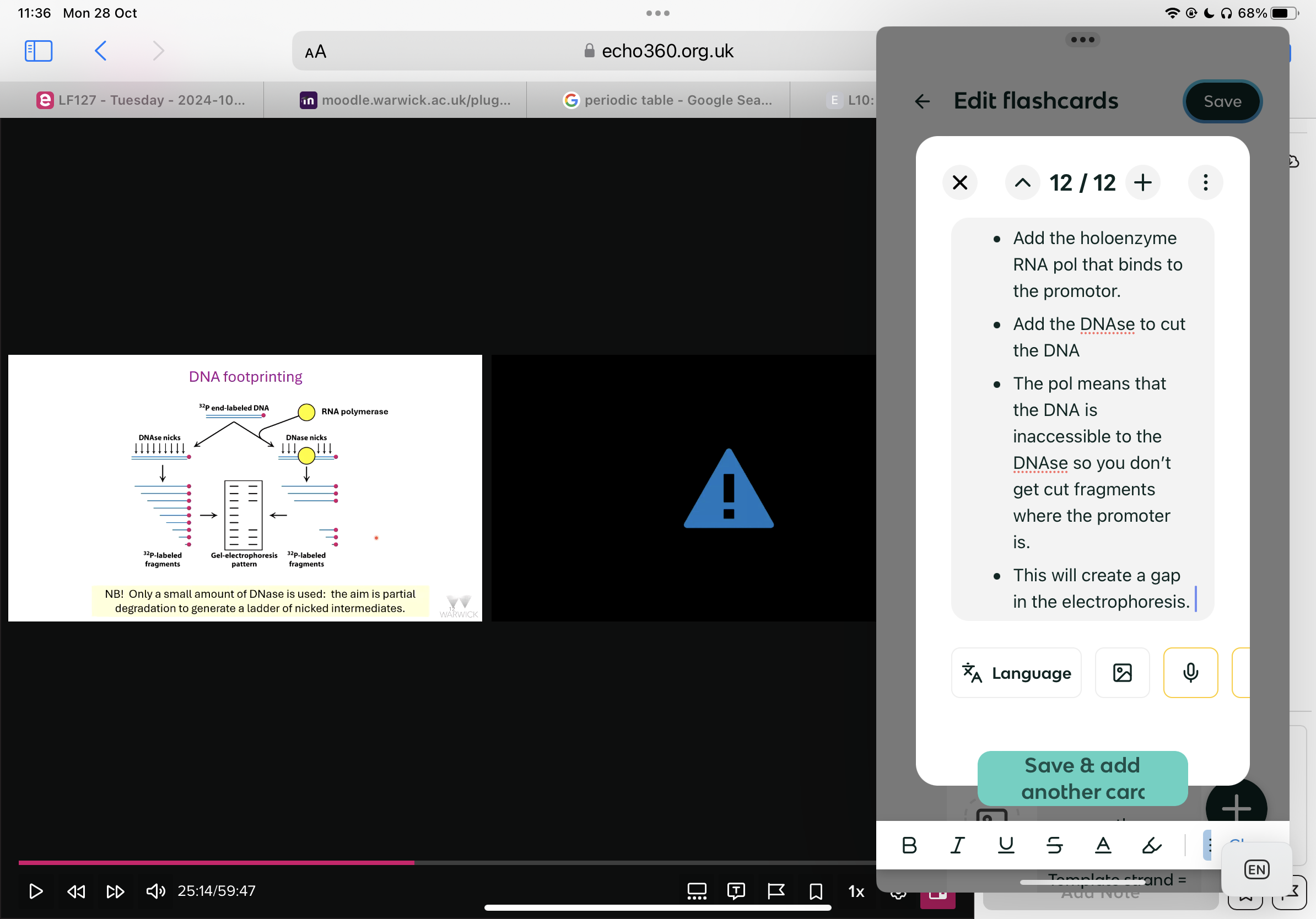
Usually two regions are protected by the RNA polymerase as it makes two contacts with the promoter. Where?
One around -10 bp from the start of the transcription and one around -35 bp from the start of transcription. The promoter sequences are asymmetric meaning pol can only bind and work in the correct direction.
New nucleotides are always added at which end?
Which of the two strands is copied?
The antisense strand from the 5’ to the 3’ meaning they are added at the 3’ end.
Initiation is stage one of transcription in bacteria:
The core RNA pol binds to DNA and can slide
Sigma subunit binds to core pol and directs it to promoter regions
There are multiple E.coli sigma factors. What is the most common one and how many genes does it control?
RpoD (RNA pol subunit D) controls roughly 1000 genes involved in growth/housekeeping
What happens next in transcription?
The pol pulls downstream DNA towards itself scrunching the DNA into supercoils until success occurs and -10 region is opened and pol can now separate the strands. This converts the closed promoter complex to an open promoter complex (doesn’t need ATP). Topoisomerases solve the problem of supercoiling.
Then 12 to 15 bp are unwound from within the -10 region to position +2 or +3 and the transcriptional site is exposed. RNA pol now…
Makes an RNA copy from the template strand using base pairing rules. Unlike DNA pol, RNA pol does not require a primer. After about 10 nucleotides of RNA synthesis, the sigma factor is exposed and disengages. The RNA pol can now elongate the new RNA.
Rate of elongation (50 nucleotides per second) is slow compared to…
DNA pol
RNA pol can edit its mistakes. How often does it make them?
One mistake every 10^4 - 10^5 nucleotides even with the proof reading functions.
There are two mechanisms for the termination of transcription in bacteria. Describe the Rho (p)-independent:
DNA encodes stop signals for transcription. Eg. A palindromic GC rich sequence followed by a T rich sequence. This is because a base paired stem can be formed where a loop forms and the only thing holding the RNA is a few weak AU base pairs.
The other is the p-dependant termination. Describe this:
P protein is a hexameric helicase that binds a C rich G poor sequence in the RNA and it uses its helicase acitvity to chase RNA pol. When it catches it, it disrupts the DNA:RNA hybrid helix, releasing the RNA.
What is rifampicin?
An inhibitor of prokaryotic transcription. It inhibits RNA pol by binding tightly to the RNA exit channel. It therefore affects initiation
Why doesn’t RNA pol need ATP to open the DNA strands?
RNA pol bends the DNA duplex that allows the duplex to be opened more easily.