Section C - Enzymes
1/129
Earn XP
Description and Tags
Dr. Li, 9/9 - 9/11
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
130 Terms
What three things define life?
Metabolism
Growth
Reproduction
What does it mean that the cell is the fundamental unit of life?
The simplest unit capable of independent existence is a cell; all organisms are made of cells
What is the hierarchy of life?
Molecules
Organelles
Cells
Tissues
Organs
Organisms
Populations
Communities
Exosystems
Biosphere
What is the average cell composition (dry weight)?
50% Protein*
15% Carbohydrates
15% Nucleic Acids
10% Lipid
10% Miscellaneous
* Almost all enzymes are proteins!!!
How do rate accelerations by enzymes compare to that of synthetic or inorganic catalysts?
Enzymes create faster rate accelerations than synthetic or inorganic catalysts
How do enzymes increase reaction rates?
Does this affect the equilibria of reactions?
Increase reactions rates by lowering activation barriers
Does NOT affect the equilibria of reactions
What are enzymes?
Which molecular do group do they usually fall under?
Enzymes are catalysts that increase reaction rates without being consumed
Most enzymes are globular proteins, but some can be RNA (ribozymes and rRNA)
Why are biological catalysts (like enzymes) the typical catalysts used by the body?
Great reaction specificity (avoids side products; enzymes can choose pathways that create more desirable products)
Milder reaction conditions (pH~7, 37˚C, stays near ideal conditions in cells)
Higher reaction rates (for quick access by the body)
Capacity for regulation (controls biological pathways; many different pathways for decomposition)
What are the two groups of enzymes?
Inorganic — Iron (Haber Process), Vanadium (V) Oxide (production of sulphuric acid), and Manganese (IV) Oxide
Organic — Digestive Enzymes and Metabolic Enzymes
What are the basic characteristics of enzymes?
Highly efficient (faster rxn rate) and specific
Lowers free energy of activation (ΔG‡)
Reduces time needed to reach equilibrium
Does NOT change equilibrium constant (Keq)
Does NOT change equilibrium concentrations
Does NOT change the free energy change for the reaction (ΔG)
What is the equation for the equilibrium constant (Keq) when a Substrate binds to an Enzyme at the active site?
Keq = [P] / [S], where:
(substrate) S ⇋ P (product)
What is the Ground State of a reaction?
Transition State?
Ground Slate — starting point for either the forward or reverse reaction
Transition State (‡) — point at which decay to substrate or product are equally likely
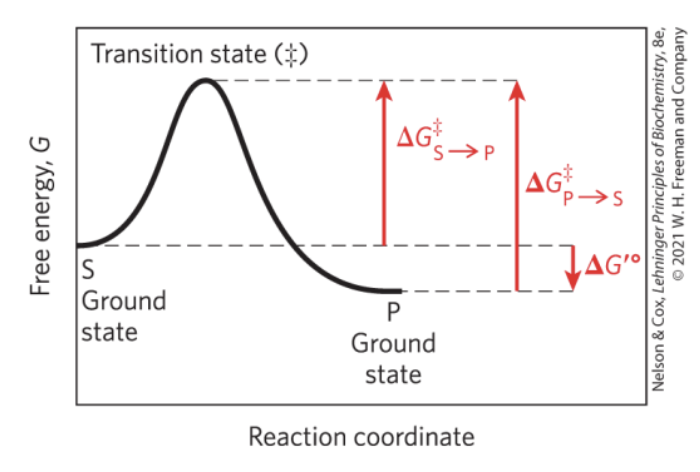
What is ∆G′° on a Reaction Coordinate Diagram?
∆G‡?
∆G′° — biochemical standard free-energy change; the standard free-energy change at pH 7.0
∆G‡ — activation energy; difference between the ground state energy level and the transition state energy level
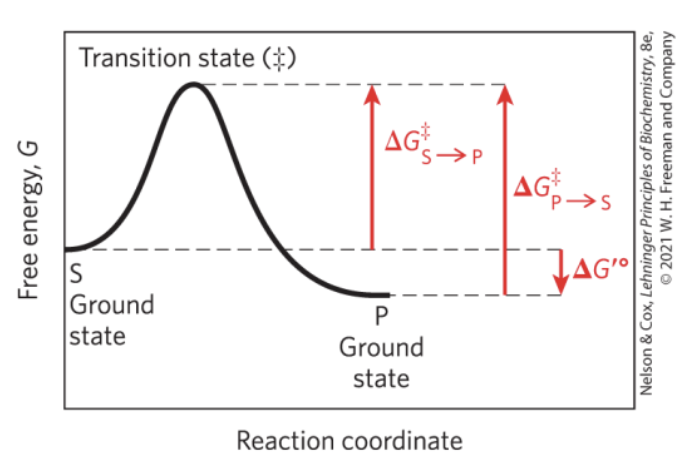
What happens to Transition State Energy (activation energy ∆G‡) when there is no enzyme present?
When there is an enzyme present?
Transition state energy is HIGH when there is no enzyme present
Transition state energy is LOW when there IS an enzyme present
What are the seven different classes of enzymes?
How are they classified?
Classified based on the type of reaction catalyzed
Oxidoreductase
Transferases
Hydrolases
Lyases
Isomerases
Ligases
Translocases
What type of reaction is catalyzed by Oxidoreductases?
Transfer of electrons (hydride ions or H atoms)
What type of reaction is catalyzed by Transferases?
Molecular group transfer
What type of reaction is catalyzed by Hydrolases?
Hydrolysis (transfer of functional groups to water)
What type of reaction is catalyzed by Lyases?
Cleavage of C—C, C—O, C—N, or other bonds by elimination, leaving double bonds or rings
Also can be the addition of groups to double bonds
What type of reaction is catalyzed by Isomerases?
Transfer of groups within molecules to yield isomeric forms
What type of reaction is catalyzed by Ligases?
Formation of C—C, C—S, C—O, and C—N bonds by condensation reactions coupled to cleavage of ATP or similar cofactor
What type of reaction is catalyzed by Translocases?
Movement of molecules or ions across membranes or their separation within membranes
What name aspects exist for each enzyme?
Each enzyme has a four-part Enzyme Commission number (E.C. number)
Each enzyme has a systematic name
Most enzymes have trivial names as well
What are Simple Enzymes?
Complex Enzymes?
Simple Enzymes — enzymes composed entirely of protein
Complex Enzymes — enzymes composed of protein + a cofactor
What two names can be used for the cofactor in a complex enzyme?
When are each of these names used?
Prosthetic Group — used when the cofactor is tightly or covalently bound
Coenzymes — used when the cofactor is non-covalently bound
What is the name for the complete complex enzyme, protein, and cofactor?
What is the name for the protein component of this complex?
Holoenzyme
Protein component is the Apoenzyme
What two groups usually function as enzyme cofactors?
What types of enzyme cofactors are they?
Vitamins and Metals can function as enzyme cofactors
Vitamins are usually coenzymes, while Metal ions are prosthetic groups
What are the two main biological and clinical significances of Enzymes?
Diagnosis
Treatment of Diseases
What does it mean when Enzymes are said to be highly specific?
Each enzyme catalyzes only ONE chemical reaction or a few closely related reactions
Where do Enzymatic reactions occur?
How is this difference than a reaction brought about using ligands?
Enzymatic reactions occur at Active Sites (specialized pockets)
These reactions occur at the Active Site, unlike in ligand-binding sites which do not have the reaction occur in that specific site
What is the Active Site of an Enzyme?
Specific region of the enzyme where the substrates bind to and where the actual reaction occurs
What are the various properties of Active Sites?
Take up a relatively small part of total enzyme volume
3D entity formed by different parts of the linear AA sequence
Is a cleft / crevice
Bound to substrates via weak, reversible attractions (HP interactions, Ionic interactions, and H-bonds)
Displays specificity of binding (depends on arrangement of atoms in the AS)
Will have a complementary shape to that of the substrate(s) (Lock and Key Model)
What is the Lock-and-Key Model?
Induced Fit Model?
Lock-and-Key — substrate fits into the AS of enzyme without needing any change or shaping
Induced Fit — enzyme AS changes shape in order to induce a fit with the substrate
Of the three states that exist during a reaction, which does an Enzyme favor most?
Why?
What level of activation energy do Enzymes favor most?
Enzymes favor the Transition State and LOWER Activation Energy
Enzymes favor the Transition State because full complement only occurs once the transition state is reached
How do enzymes lower the ΔG‡ of a reaction?
Enzymes bind best to transition states; this means that when the transition state exists, enzymes will bind tightly and effectively lower the activation energy barrier
What idea was proposed by Linus Pauling in 1946?
What were the various components of this idea?
Enzymes bind transition states best
Enzyme AS are complementary to the TS of the reaction
Enzymes bind TS better than substrates
Stronger interactions with the TS compared to the ground state (GS) lower the activation barrier
This is largely a ΔH‡ effect (enthalpic interactions, i.e. interactions)
What two concepts are used to explain the catalytic power of Enzymes?
Enzymes bind most tightly to TS of the cat. reaction (lowers activation barrier)
Enzyme AS are organized by evolution to facilitate multiple mechanisms of chemical catalysis simultaneously
What is the main role of Binding Energy in Catalysis?
Weak binding interactions between the Enzymes and the Substrate drive enzymatic catalysis (not too strong, not too weak)
Must be strong enough to overcome the Barrier to Reaction
What components contribute to the Barrier to Reaction in enzymatic catalysis?
Entropy of molecules in solution
Solvation Shells of H-bonded water around most biomolecules in aqueous solution
Distortion of substrates that has to occur in reactions
Need for proper alignment of fxn groups on the enzyme
What are the three main catalytic mechanisms?
Do enzymes use only one mechanism or can they use multiple?
Acid-Base Catalysis — give and take protons
Covalent Catalysis — change reaction paths
Metal Ion Catalysts — use redox cofactors; pKa shift
Enzymes can use one or combinations of these mechanisms
What is the general idea behind Acid-Base Catalysis?
What are the two specific types of Acid-Base Catalysis?
Does this type of mechanism occur in most enzymes?
Protons are transferred between an enzyme and a substrate / intermediate
Two types are Specific AB Cat. and General AB Cat.
Yes, this mechanism occurs in most enzymes
What is Specific Acid-Base Catalysis?
General Acid-Base Catalysis?
Specific — uses ONLY the H+ or OH- ions present in water
General — mediated by weak acids or bases other than water
What is the general idea behind Covalent Catalysis?
When does this occur?
Transient covalent bonds are formed between the enzyme and the substrate
Only occurs when the new pathway (w/enzyme) has a lower activation energy than the uncatalyzed pathway (ALL new steps must be faster)
Which catalytic mechanisms does Chymotrypsin use?
Combination of Acid-Base Catalysis and Covalent Catalysis
What is the general idea behind Metal Ion Catalysis?
How many known enzymes require more than one ion for catalytic activity?
Metal ions help orient the substrate for the rxn, allowing for stabilization of charged reaction TS
Also help mediate Redox reactions by allowing for reversible changes in the metal ion oxidation states
About 1/3 of all known enzymes require more than one metal ion for catalytic activity
What is a Protease?
Give an example of a Protease. How does it carry out its function?
Enzyme that catalyzes the hydrolytic cleavage of peptide bonds
Example of a Protease includes Bovine Pancreatic Chymotrypsin (acylation and deacylation of a Ser residue)
How are Chymotrypsin-Catalyzed reactions dependent on pH?
What changes causes each transition?
What happens due to each transition?
Optimal activity at pH 8 (His57 unprotonated, Ile16 protonated)
Transition just above pH 7 happens due to changes in kcat (His57 is protonated)
Transition just above pH 8.5 happens due to changes in 1/Km (Alpha-amino group of Ile16 is ionized)
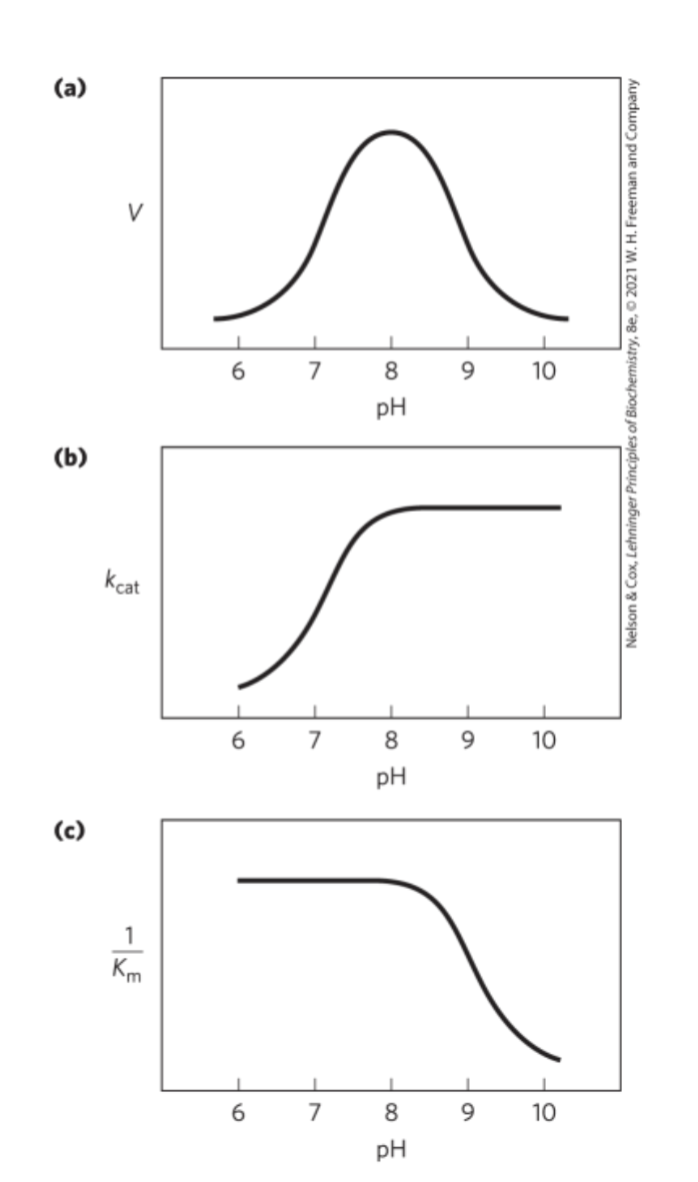
What is the main function of Chymotrypsin?
What specific reactions does it carry out?
Chymotrypsin is one of several proteases that cuts peptides at specific locations on the peptide backbone
Carries out cleavage reactions of peptide bonds adjacent to Aromatic AAs
What determines the substrate specificity of an Enzyme?
Substrate specificity of an enzyme is determined by the structure of the substrate-binding site
Which Amino Acids have Aromatic R Groups in their structure?
Positively charged R groups?
Aromatic R Groups — Phenylalanine (F), Tyrosine (Y), and Tryptophan (W)
Positively Charged R Groups — Lysine (K), Arginine (R), and Histidine (H)
What is a Serine Protease?
How do they contribute to the Chymotrypsin Reaction?
Serine Protease — proteases with a Ser residue that acts as a nucleophile
The nucleophile that acts in the Acylation phase of the reaction is the Oxygen of Ser195
What is a Catalytic Triad?
How does this relate to the Chymotrypsin Reaction?
Catalytic Triad — hydrogen bonding network
In the Chymotrypsin Reaction, there is a catalytic triad made up of Ser195, His57, and Asp102
What is Step 1 of the Chymotrypsin Mechanism?
Step 1 is Substrate Binding
Substrate binding allows the side chain of residue adjacent to the target peptide bond to nestle into a hydrophobic pocket on the enzyme, allowing room for the cleavage attack by the Catalytic Triad (Ser195, His57, and Asp102)
What is a Scissile Bond?
Scissile Bond is the specific bond that is susceptible to being cleaved in a reaction
What is Step 2 of the Chymotrypsin Mechanism?
Step 2 is Nucleophilic Attack
Oxygen of the Ser195 acts as a nucleophile and attacks the carbonyl group of the peptide, forming a tetrahedral acyl-enzyme
Short-lived negative charge on the carbonyl oxygen is than stabilized by H-bonding in the oxyanion hole (short-lived acylation intermediate)
What is Step 3 of the Chymotrypsin Mechanism?
Step 3 is Substrate Cleavage (via Covalent and AB Catalysis using the Catalytic Triad)
Instability of the short-lived negative charge on the carbonyl carbon leads to collapse of the tetrahedral intermediate
Double bond with carbon reforms, displacing and breaking the bond between carbon and the amino group of the peptide bond
Amino leaving group is then protonated by His57, leading to its displacement
What is Step 4 of the Chymotrypsin Mechanism?
Step 4 is Water Coming In
Incoming water molecule is deprotonated by General Base Catalysis, creating a strongly nucleophilic OH-
OH- then attacks the ester linkage of the Acyl-Enzyme, generating a second tetrahedral intermediate
Oxygen takes on a negative charge again and is located in the oxyanion hole
What is Step 5 in the Chymotrypsin Mechanism?
Step 5 is Water Attacks
Second tetrahedral intermediate collapses and forms the second product (carboxylate ion) that then displaces Ser195
What is Step 6 in the Chymotrypsin Mechanism?
Step 6 is Break Off from the Enzyme
Second product (carboxylate ion) dissociates from the AS, regenerating a free enzyme
What is Step 7 in the Chymotrypsin Mechanism?
Step 7 is Product Dissociation
Product is the smaller peptide fragments resulted from the cleavage of the peptide bond
What are the steps of the Chymotrypsin Mechanism?
Substrate Binding — substrate binds, cleavage of Scissile Bond by Catalytic Triad
Nucleophilic Attack — Ser195 nucleophilic alkoxide attacks, forms tetrahedral intermediate and short-lived negative charge
Substrate Cleavage — collapse of tetrahedral intermediate, breakage of peptide bond, and displacement of amino leaving group
Water Comes In — incoming water generates nucleophilic hydroxide, hydroxide attacks and generates another tetrahedral intermediate, and oxygen takes on negative charge again
Water Attacks — second tetrahedral intermediate collapses, forming carboxylate ion product and displacing Ser195
Break-Off From the Enzyme — carboxylate ion product dissociates from AS, regenerates free enzyme
Product Dissociates — smaller peptide fragments dissociate
What is Kinetics?
Kinetics — study of compound reaction rates and how they change in response to changes in experimental parameters
What factors change the rate of enzymatic reactions?
Enzyme
Substrate
Effectors
Temperature
What is the rate equation for enzyme kinetics?
What factors can affect this reaction rate?
Rate Equation: V = k[S], where:
V = Velocity
k = rate constant
[S] = substrate concentration
Rate can be affected by:
pH
Temperature
Enzyme Conc.
Substrate Conc.
How does temperature affect the rate of enzymatic reaction?
Enzyme concentration?
There is an optimal temperature for enzymatic reactions
As enzyme concentration goes up, the rate at which the reaction occurs goes up as well (directly linear)
What is the relationship between the Michaelis Constant (Km) and Binding Affinity?
Inverse relationship (Higher Km = Lower Binding Affinity)
When does V0 start to plateau on a graph of Substrate Conc. vs. Initial Velocity?
V0 starts to plateau the closer it gets to Vmax
What equation is used to determine V0 in Michaelis-Menten Kinetics?
V0 = (Vmax)[S] / (Km) + [S]
What is the equation for k1 in Michaelis-Menten Kinetics?
k2?
k3?
What is k3 equivalent to?
k1 = [ES] / [E][S]
k2 = [E][S] / [ES]
k3 = kcat = [E][P] / [ES] or Vmax / [Et], where [Et] is conc. of total enzyme
What is the equation for V0 in Michaelis-Menten Kinetics?
Vmax?
V0 = kcat[ES]
Vmax = kcat[Et], where
Et = total enzyme conc.
What aspect of the Time vs. Product Conc. [P] curve is used to determine Initial Velocity of the Enzyme reaction?
What does this aspect represent?
Initial Velocity (V0) is taken as the tangent line of the Time vs. [P] curve at time = 0
Represents that Enzyme-Catalyzed Reactions are Steady State Equations
What is important to remember about [S] at the start of the enzyme-catalyzed reaction?
What happens as time goes on in the reaction?
[S] at the start of the EC reaction is constant
As time goes on, [S] decreases as it is used up by Enzymes
What are the relative concentrations of the Enzyme and Substrate in Michaelis-Menten Kinetics?
[S] >>> [E]
The amount of substrate bound by the enzyme at a time is small
What does it mean that the Michaelis-Menten Equation follows a Steady-State Assumption?
[ES] does NOT change with time (formation of [ES] = breakdown of [ES])
Rate of synthesis = Rate of degradation
Which reaction rates do we use when calculating and analyzing Enzyme-Catalyzed reactions that follow Michaelis-Menten Kinetics?
We only use initial reaction rates (rate from when the enzyme and substrate are initially mixed)
At this time, [P] is VERY small and the rate of the back reaction can be ignored
What is the Michaelis-Menten Equation?
V = (kcat)[Et][S] / Km + [S], or
V = (Vmax)[S] / Km + [S]
What does kcat represent in the MM Equation?
Km?
kcat — Turnover number; how many S molecules one E molecule can convert per second
Km — Michaelis Constant; approx. measure of a Substrates affinity for an Enzyme; [S] value in which Vmax is observed
During a Steady State Assumption, when does the Vmax occur?
What is it dependent upon?
Vmax occurs when all of the Enzyme is in the ES complex
Dependent on the breakdown of that complex, k[ES]
What happens to V0 when we have high [S]?
Which equation do we use?
Velocity is no longer proportional to [S] at high [S]
V0 = kcat / Km
What is the Hyperbolic Form of the MM Equation?
Linear Form?
Hyperbolic: V0 = (Vmax)[S] / (Km + [S])
Linear: (1 / V0) = (Km + [S]) / (Vmax)[S]
What type of graph do we observe when we plot 1/V0 vs. 1/[S] with MM Kinetics?
What does the Slope represent?
X-intercept?
Y-intercept?
We generate a linear graph (straight line)
Slope = Km / Vmax
X-int = -1 / Km
Y-int = 1 / Vmax
What limits enzyme efficiency?
How can we gain efficiency during Enzyme-Catalyzed reactions?
Specificity stemming from active site diffusion limits the efficiency
We can gain efficiency by increasing V0 or Substrate Affinity
What are Enzyme Inhibitors?
What two classes of enzyme inhibitors exist?
Molecules that interfere with Catalysis by slowing or halting enzymatic reactions
Two classes are (1) Reversible and (2) Irreversible
What are the four different types of Reversible Enzyme Inhibition?
Competitive Inhibition
Uncompetitive Inhibition
Mixed Inhibition
Noncompetitive Inhibition
What is a Competitive Inhibitor?
Noncompetitive Inhibitor?
What types of inhibition do these inhibitors carry out?
Competitive — competes with the substrate for the AS of an enzyme
Noncompetitive — binds at a site distinct from the substrate AS; binds specifically to either the E or the ES complex
Both inhibitors carry out Reversible Inhibition
What are Irreversible Inhibitors?
Inhibitors that bind covalently to or destroy a functional group on an Enzyme essential for the enzyme’s activity
Can also form a highly stable noncovalent association
Covalent Bond destroys a necessary functional group
What is the main function of Methotrexate as an Inhibitor of Thymidylate Synthase?
What type of inhibition is this?
Inhibits replication of cancer cells and is used for Chemotherapy / as an immunosuppressive drug
Competitive Inhibition
What do Competitive Inhibitors do to:
Substrate Binding Affinity
Km
Vmax
The [S] vs. V0 curve
Substrate Binding Affinity — decreases
Km — increases
Vmax — no effect
[S] vs. V0 curve — shifts to the right
What is the MM Equation when dealing with Competitive Inhibitors?
What is the x-int?
Y-int?
Slope?
MM Equation: 1 / V0 = {(αKm) / (Vmax)[S]} + 1/Vmax
X-int: -1 / αKm
Y-int: 1 / Vmax
Slope: αKm / Vmax
Where will all the lines intersect when there is a Competitive Inhibitor?
Noncompetitive Inhibitor?
Lines intersect on the y-axis for Competitive Inhibition
Lines intersect on the x-axis for Noncompetitive Inhibition
What do Noncompetitive Inhibitors do to:
Substrate Binding Affinity
Km
Vmax
Catalytic Power of the Enzyme
Substrate Binding Affinity — stays the same
Km — no effect
Vmax — decreases
Catalytic Power — decreases (effective [E] decreases)
What are Regulatory Enzymes?
What do they allow?
Regulatory Enzymes — increases or decreases catalytic activity in response to certain signals
Allows the cell to meet changing needs for energy and biomolecules
What are the different ways in which the activity of a Regulatory Enzyme can be modulated?
Via allosteric enzymes
Reversible covalent modification
Binding of separate regulatory proteins
Removal of peptide segments via proteolytic cleavage
What are Allosteric Enzymes?
Enzymes that function through reversible noncovalent binding of regulatory compounds (Allosteric Modulators / Effectors)
What types of molecules typically act as Allosteric Modulators or Allosteric Effectors?
Metabolites or Cofactors
slide 69
what this mean/??????
What is the main premise of the Concerted Model?
There exists a Relaxed / High Activity State and a Tense / Low Activity State
Both states exist in equilibrium, but at different %’s of each state
What is the main premise of the Sequential Model?
Each subunit of an Allosteric Enzyme converts individually
Once a subunit binds, it changes the conformation of the other subunits so as to make binding easier for them
How do Allosteric Enzymes relate to MM Kinetics?
What types of curves do they create when plotting [S] vs. V0?
The kinetic properties of Allosteric Enzymes DIVERGE from MM Kinetics
Allosteric Enzymes produce sigmoidal curves instead of the normal MM Hyperbolic curves
What does [S]0.5 or K0.5 represent?
[S] that gives ½ Vmax