Gene Regulation and Development
1/10
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
11 Terms
gene expression
Cells can turn genes on or off when needed
This depends on environmental or internal signals
“On” = gene is transcribed
“Off” = gene is not transcribed
This helps with cell specialization (like how skin and nerve cells are different)
bacterial gene expression
operons are a group of genes that can be turned on or off, they have 3 parts
promoter: where RNA polymerase attaches
operator: on/off switch
genes: code for related enzymes in pathway
operons are repressible or inducible
repressible (on to off): transciption is usually on but can be stopped
inducible(off to on): transcription is usually off, but can be induced
bacterial gene expression
regulatory gene → makes a repressor protein that binds to the operator to block RNA polymerase from transcribing the gene
always expressed, but at low levels
binding a repressor to an operator is reversible

review on allosteric enzymes
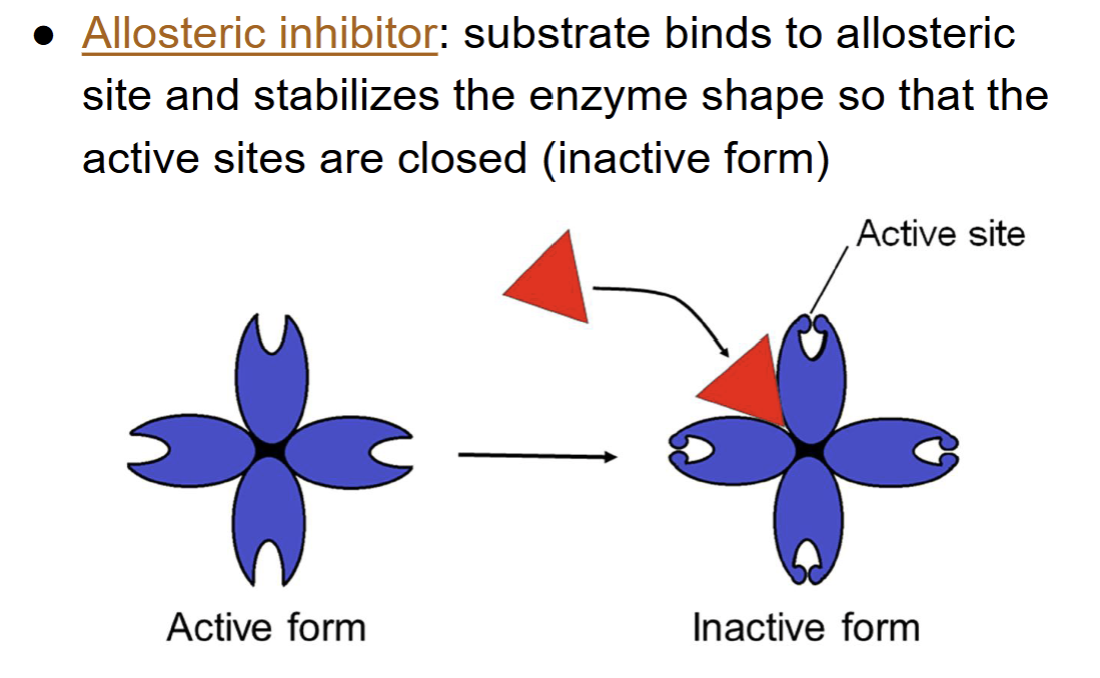
allosteric activator → a substrate binds to the allosteric site and changes the enzyme’s shape to keep the active site open and working properly
allosteric inhibitor → a substrate binds to the allosteric site and changes the enzyme’s shape so the active site closes, keeping the enzyme inactive
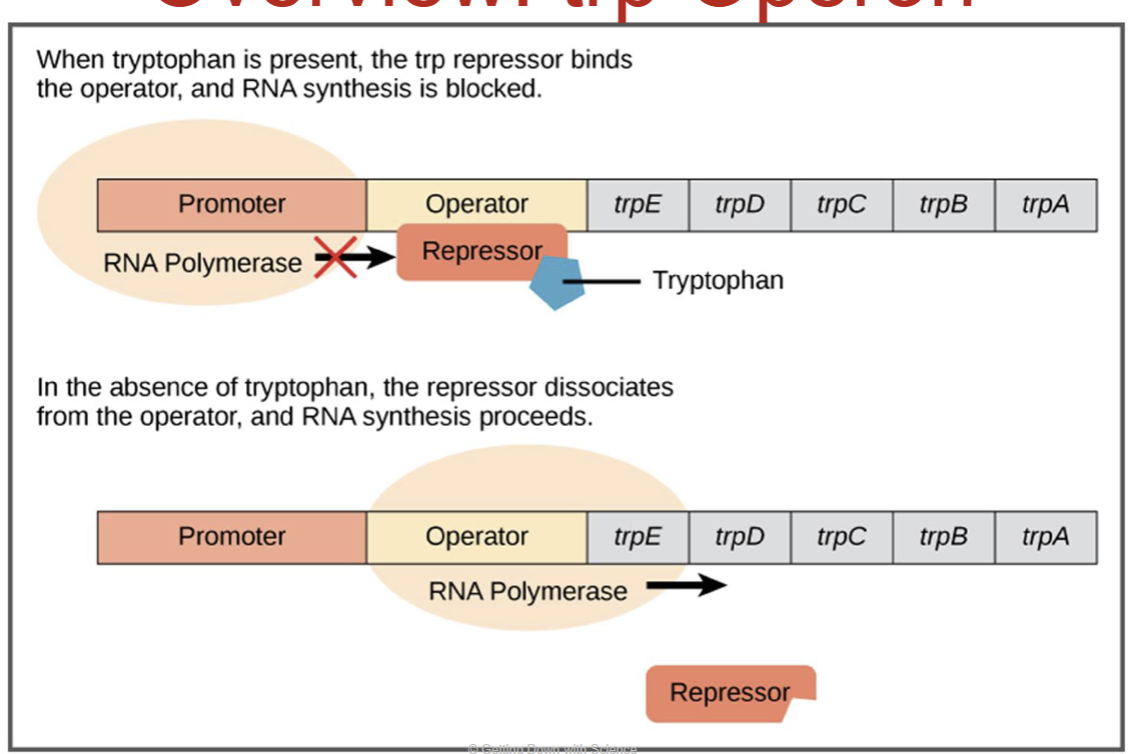
repressible operons - trp operon
trp operon helps bacteria make tryptophan
a repressible operon, so it’s usually on
A repressor protein can turn it off
repressor is inactive by itself
when tryptophan levels are high, tryptophan binds to the repressor
this activates the repressor, which then binds to the operator
This blocks transcription, so no more tryptophan is made until it’s needed again
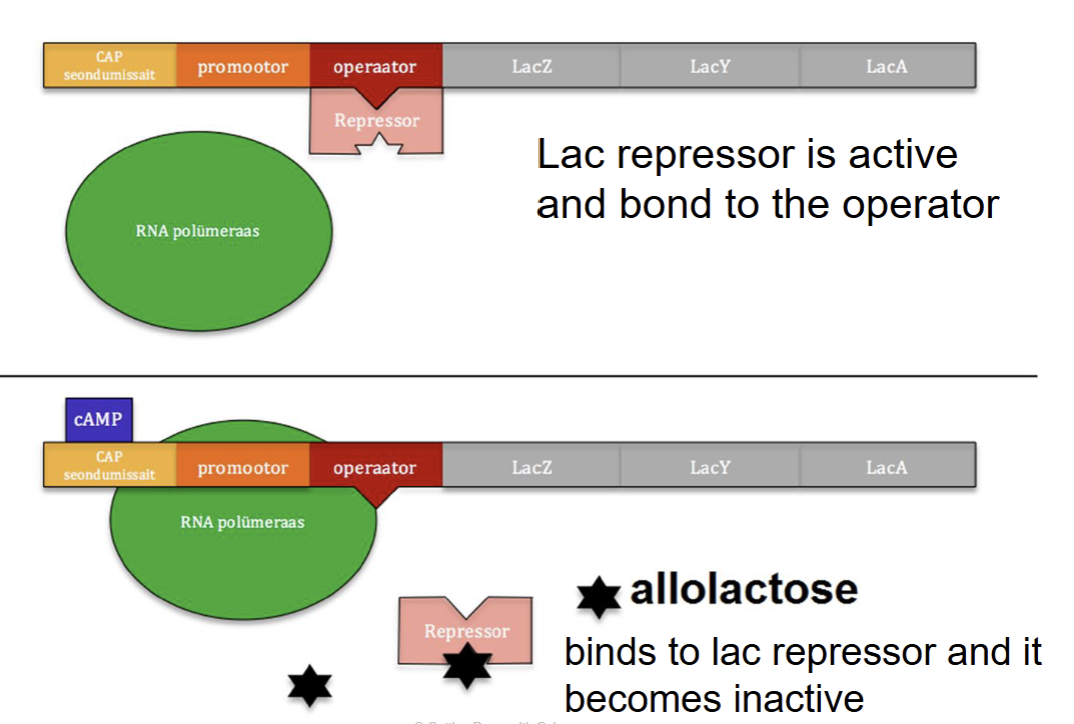
inducible operons - lac operon
controls making lactase, which breaks down lactose
It’s an inducible operon, so it’s usually off
normally bound to the operator, blocking transcription
lactose (allolactose) is present, it acts as an inducer
allolactose binds to the repressor, turning it inactive
The repressor falls off, and the genes can now be transcribed to make lactase
eukaryotic gene expression pt.1
eukaryotic gene expression is regulated at different stages
chromatin structure → if DNA is tightly wrapped its less accessible for transcription
histone acetylation → adds acetyl groups to histones → looses DNA
DNA methylation → adds methyl groups to DNA → chromatin condense
epigenetic inheritance
chromatin changes don’t change the DNA sequence, but they can be passed down
these changes can be reversed, unlike mutations
explain why identical twins can have different traits or health conditions
eukaryotic gene expression pt.2
transcription initiation
chromatin is loosened, transcription factors can bind to special control elements (non-coding DNA parts).
these can increase or decrease gene expression depending on whether activators or repressors bind to them.
eukaryotic gene expression pt.3
RNA processing
alternative splicing of pre-mRNA
translation initiation
translation can be activated or repressed by initiation factors
microRNAs and small interfering RNAs can bind to
mRNA and degrade it or block translation
eukaryotic development
during embryonic development, cell division and
cell differentiation occurs
cells become specialized in their structure and function
morphogenesis → physical process that gives an organism its shape
how do cells differentiate during early development?
cytoplasmic determinants → substances in the mom egg that influence cells
induction → cell-to-cell signals that can cause a change in green expression
both influence pattern formation
“body plan” for the organism
homeotic genes map out body structures
hox genes are homeotic genes; they are known for their role in specifying the identity of body segments during embryonic development.
apoptosis → programmed cell death
allows structures to take their form
ex: if apoptosis did not occur during the development of human hands and feet, we would be born with webbed fingers and toes