hmb265 2nd half
1/93
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
94 Terms
Lecture 16: Transposable Elements
do genes exist in fixed places in the chromosome?
no. one woman revolutionalized…. with only classical genetics and a light microscope. what a woman
Barbara McClintock
late 1940s
discovered movement of small segments of DNA transposable elements
used maize
one train of maize showed chromosome 9 broke often at a particular site
involve Ds element and Ac element
molecular structure of DNA was unknown
using classical genetics and a light microscope
transposable elements
movement of small segments of DNA
Ds element
dissociation factor
site of the break
non autonomous element - cant mobilize itself
incomplete mutated form of Ac element
Ac element
unlinked genetic factor necessary to be present to activate the breakage at the Ds locus
autonomous element - can moblilize itself and other mobile elements in the same family
can become a Ds element (when it cant jump back out again)
inverted repeats at the end, encodes a single gene which encodes a transposase protein
chromosome 9
controls genes that control several phenotypes of maize kernels;
pt traits: pigmented/colorless, plump/shrunken, nonshiny/waxy
transpoable/unstable alleles can lead to novel/odd phenotypes
spotted phenotype: independent exicsion of Ds alleles
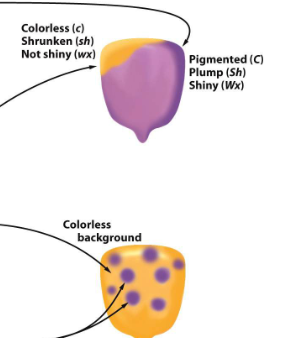
unstable phenotype
characteristic of mobile/transposable elements
*add more detail/definiton
mechanism of transposition in corn
transposase
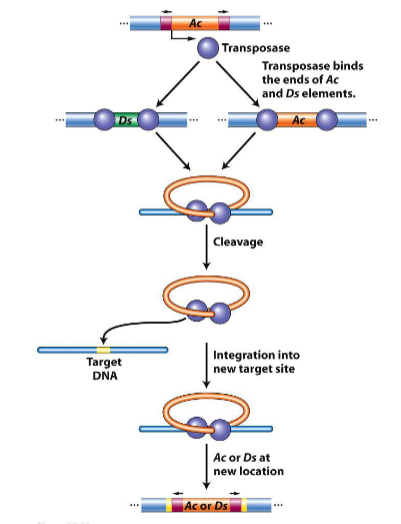
insertion of element
cuts with staggered ends (like restriction enzymes), inserts transposable elements between staggered ends→cell repairs ends
net result: flanking sequences, direct repeats reflecting a repeat of the target site dna generated by processes of dna repair
T or F? Transposable elements are common.
True.
Found in all kinds of organisms; bacteria, Drosophila (12.5%), humans (34% of the genome)
2 main classes of transposable elements in eukaryotes
retrotransposons & DNA transposons
retrotransposons
transposable elements going through an RNA intermediate
different types; some have LTRs long terminal repeat sequences (100s of bases long; direct repeats)
similar to retroviruses; go through RNA intermediate which then gets translated into a reverse transciptase → cDNA→reinserted into genome in a different place. however they do not get packaged and leave the cell; stay moving around the genome.
DNA transposonas
smaller than retrotransposons
gene encodes a transposase in the gene
eg P element DRosophila,
recognizes inverted repeats, excies element, transposes to new genomic lovcation
original location has a gap and either a)homologous chromosome without P elemnt is repaired with no transposon or b) p element reconsitiuted in original position if sister chromatid
transposable elements in humans
LINEs, SINEs, DNA transposons
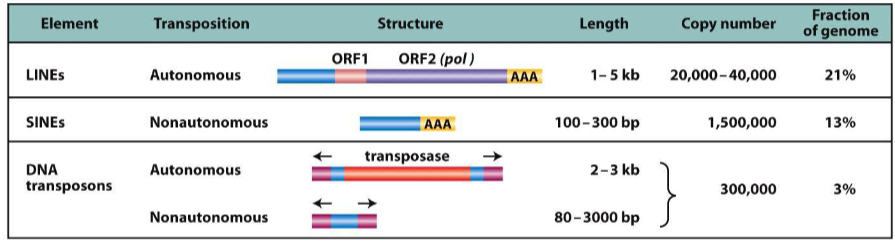
LINEs
req reverse transcriptase to mobilize
autonomous
SINEs
req reverse transcriptase to mobilize
non autonomous
how do organisms survive with so many mobile elements?
so much of our DNA is transposable elements- but we can control this mobilization
they are found in introns
often defective unable to transpose again due to mutations (lack of repeats or active transposase)
epigenetic changes in heterochromatin vs euchromatin “silencing” parts of genome make “safe havens”
but mutation can occur
chromosomal rearrangments
Transposable elements can result in unqueal lining of homologous chromosomes; alter dna
can relocate dna sections
lead to inapporpriate expression of gene product
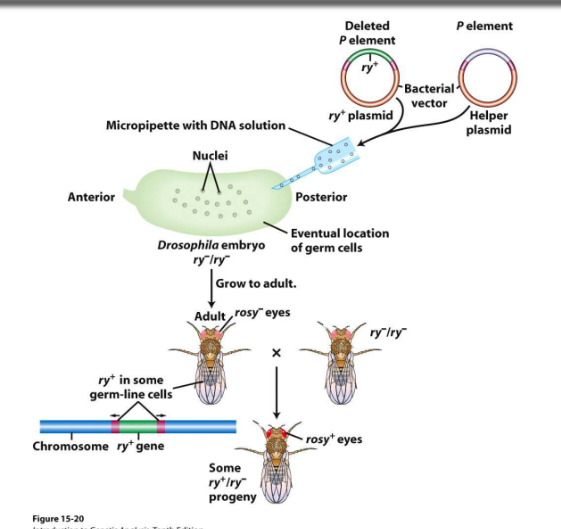
laboratory uses of TrElems
allows for the transfer of any gene of interest to make transgenic flies
replace transposase gene with marker gene and inject into drosophila embryo
subject will have rosy- phenotype, but with a successful transposition some of progeny crossed with WT will be rosy wildtype
Lecture 13: Quantitative trait loci
How to define heritability?
What is heritable in a gene?
P=G+E
phenotype is the sum of genes + environment
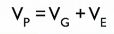
variation in phenotype = Sum of variation of genetics and variation of environment
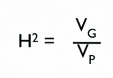
Broad Sense Heritability (H²)
the extent of phenotype variation attributable to genetic variation
H² = 1 if…
all phenotypic variation because of genetics
H² = 0 if…
all phenotypic variation because of environment
(the minimum)
H² limitation
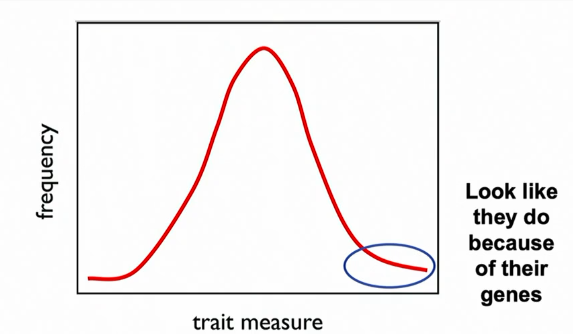
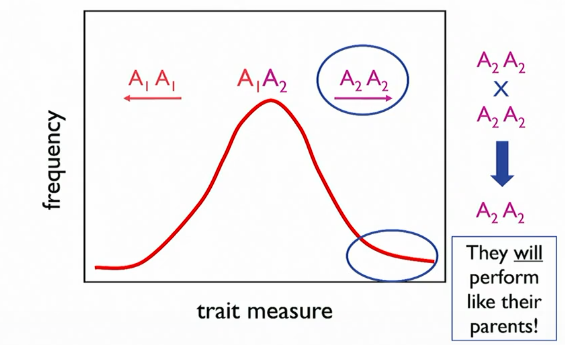
H² does not predict how progeny will perform on the basis of the phenotype of their parents
progeny=/= look like their parents
why? most variation is not transmissible from parent to offspring bc of context. offspring inherit alleles but not genomic context. interactions b/w loci are not inherited. all effects not transmitted; only additive effects
experiments for H², Vg, and Vp
measure effect of genotypes in many environment
model species, genetically identical strains
hold Vg as close to 0
twin studies
show environmental effects, where genes are “held constant”
If H² is high…
the phenotype of an individual is probably because of genotype IN THAT FAMILY (in the same population where env and stuff are relatively constant)
What H² DOESN’T tell us
phenotype of individual based on parents phenotypes, even if H² is high
what is going on in other families/populations
What H² DOES tell us
the proportion of phenotypic variability attributed to genetics for the entire population.

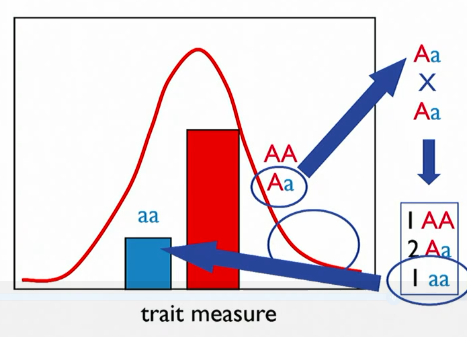
3 components of genetic variation
additive, dominance, epistatic effects

Vd
phenotype not predictable - may be homozygous or heterozygous
Vi
cant predict phenotype
context of another locus matters

Va
predictable
narrow sense heritability (h²)
extent of phenotypic variation attributable to additive genetic variation
what is commonly thought of as ”heritability”

if h² = 1…
all the phenotypic variation is attributable to additive variation
the maximum
if h² = 0…
if phenotypic variation is attributable to other genetic & environmental effects
the minimum
h² does tell us
the phenotype is predictable based on parent phenotype for that specific place, time people, family, population etc
eg. selective/artificial breeding if h² is high (eg. pick a large pig to make more large pigs)
h² doesnt tell uss
whats happening in other families
what the genes are
Lecture 14: Genetic Mapping & Complex Traits
quantitative effect
usually two or more important genes
Quantitative Trait Loci (QTL)
genes controlling the trait in a quantitative loci
regions of genome associated with variation in a complex trait
identified using genetic markers (eg SNPs), association studies, statistical methods
Step 1
cross b/w inbreds which differ at trait(s) of interest
because limited by genetic diversity present in inbreds (less recombination) → variation in phenotype due to env. causes
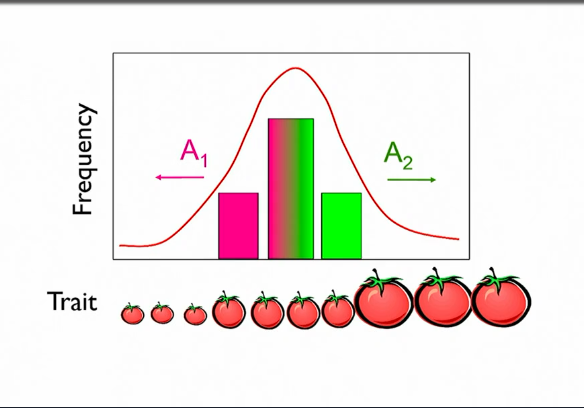
Step 2: F2 frequency distribution
does gene distribution match phenotype distribution?
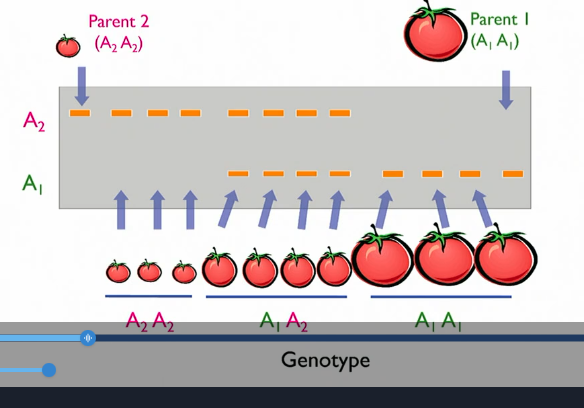
Step 3: molecular markers; genotyping
molecular markers spread out across genome at regular intervals to find markers that co-segregate with trait ()
sequence all F2
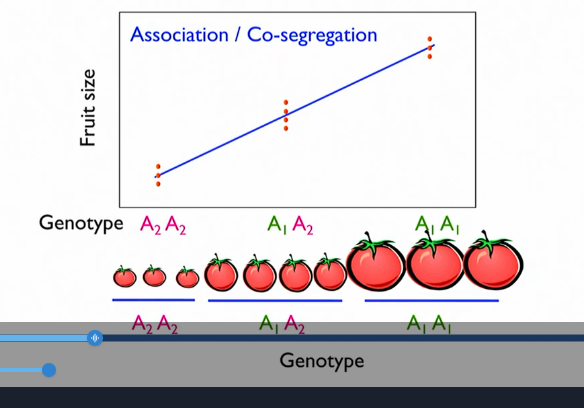
Step 4: statistics ; are markers co-segregating?
statistically significant →
associating with the trait or not
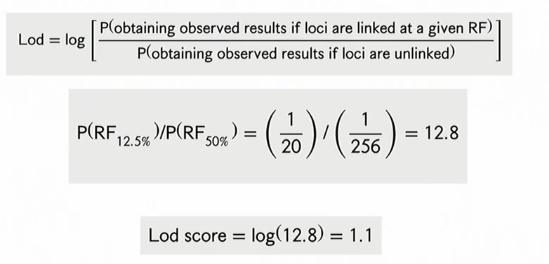
step 5: LOD score
plot the degree of association (logsrithm of oddds LOD) on a linkage map
statistical association between a marker and a trait of interest?
Odds Ratio (OR)
probability of linkage →in favor of QTL
LOD = log OR
100 → 100 odds in favor of there being a QTL

LOD score example
LOD > 3 → linked
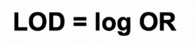
Lecture 17: Chromosome Packing & Number
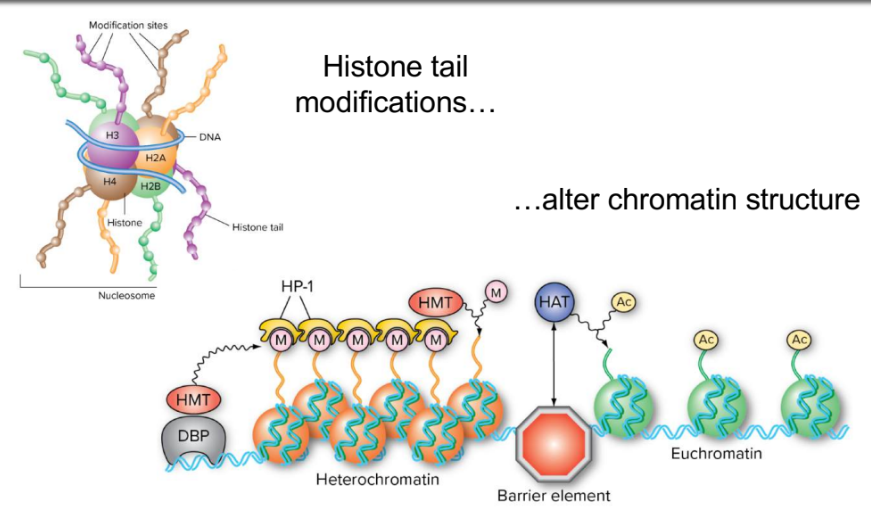
chromosome packing : nucleosome
double stranded helix wrapped around histone core, 2 sets of 4 subunits
organizational purposes
can be open or closed
accessible or inaccessible for transcription & regulation
can protect from cleaving/digestion
heterochromatin
transctriptionally inactive
extremely closed conformation
dna silenced
euchromatin
transcriptionally active
chromatin remodelling
histone tail modifications (100+ diff ways)
acetylation (by HATs)
methylation (usually of K or R) by HMT
alter chromatin structure
huge flexibility in complexity
x-chromosome inactivation
heterochromatin formation inactivates an entire x chromosome (into a Barr body)
random which one gets inactivated
early during development
2nd x is still essential until 100 cell count
inactivation is hereditary through cell division i.e. clonal
(except reactivated in germ cells)
reason—dosage compensation
still a small number of genes on barr body expressed
mechanism of x inactivation
XIST RNA coats a chromosome creates Barr body/ heterochromatin
hypoacetylation of a Lys in two histones & histone methylation
euploidy
complete sets of chromosomes
eg. diploid (2 sets of chromosomes, humans), monoploid (one set), triploidy
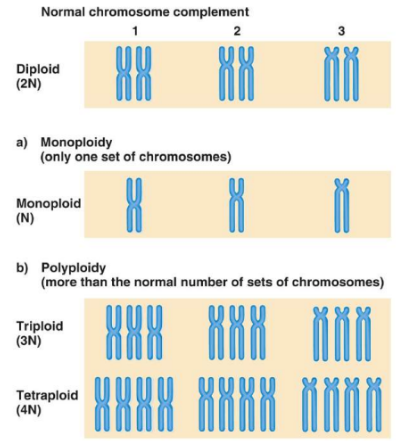
aneuploidy
loss or gain of one or more chromosomes
eg. monosomy (missing a chromosome in a pair of homologous chromosomes in chrom 3
usually caused by meiotic nondisjunction; diff distributions of chromosomes in gametes (extra or lacking chromosome)

monoploidy
common in male social insects
male bees, wasps and ants
develop gametes vua parthenogenesis
unfertilized egg into an embryo, males produce gametes via modified meiosis
can create monoploid plants uncover recessive mutations
take meisois product in anther and cause haploid cell in lab to be induced to grow as an embryo and eventually formed monoploid plantlet and observe phenotype visualizing traits directly and study mutations
polyploidy
associated with origin of new species
+ correlation with size and vigor
reason fruits are getting bigger
more polyploid (more sets of chromosomes) , bigger fruits
autopolyploids
form spontaeneously or in lab
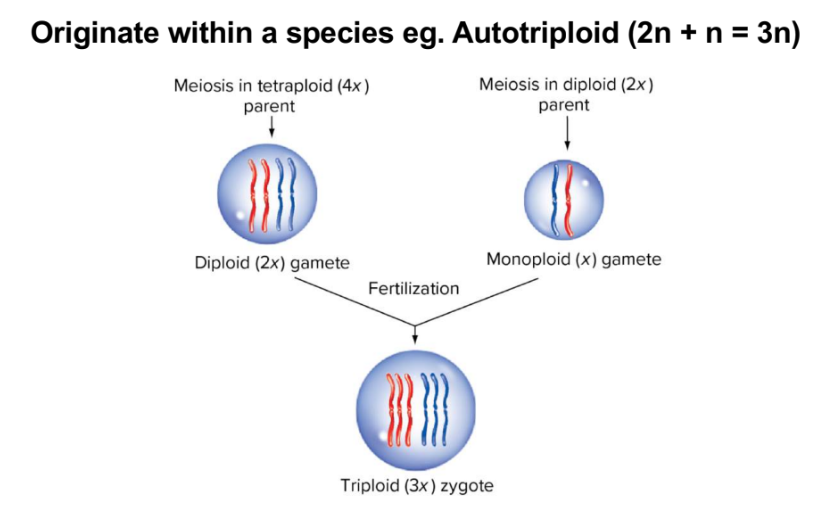
autotriploid (or other odd set)
tend to be sterile
issue pairing chromosomes in meiosis
may form unbalanced gametes
chance of complete set of balanced chromosomes is very low but possible
commercial bananas and watermelons are sterile; propogated asexually
autotetraploids
can be spontaneous doubling (2n→4n) or drug induced (colchicine)
usually source of new species because new plant self fertilizes is not distinctly tetra ploid and cant be crossed back to the diploid parent
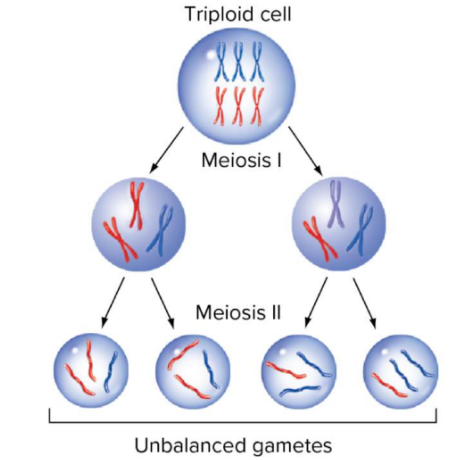
tetraploid meiosis
in tetraploidy you dont have 2 homolog chromosomes you have 4 homologous chromosmes for each different type of chromosome, when pairing up, only homologues can pair up, they have a choice of three partners to pair up with (1+2 new)
once paired with 3 possible partners, different ways of assorting”left door or right door” for one partner and the other
pairing and alignment varies
4 alleles instead of 2
all possibilities are equally likely
choices of who they pair with and which way they assort
probability of aa aa gametes for example are 2/12 or 1/6 ² because 2 parents → 1/36
unique dihybrid ratio→ 1AA:4Aa:1aa
allopolypoloids
hybrids of two closely related species where chromosomes are partially but not fully homologous
F1 sterile
homeologous
chromosomes or genes that are similar but have originated from different parental species
Homoeologs are pairs of genes that originated by speciation and were brought back together in the same genome by allopolyploidization
(fertile) amphidiploid
doubled diploid: doubling in germ cells
type of alloploid
monosomy
usually lethal in humans in utero
missing a whole chromosome
eg monosomy 21
turner syndrome (XO)
trisomy
2n+1
trisomy 21 down syndrome
smaller size →less bad result (eg down syndrome chromosome 21 is smaller than other maybe more lethal disorders from other chroomosomes)
down syndrome
chromosome 21 has 3 chromosomes instead of 2 like all the other ones
bivalent chromosomes
i think just homologous chromosomes
trisomies XXX
less bad
XYY: Usually fertile – X pairs with one Y; other Y does not
pair and is not transmitted to gametes
i.e. X or Y gametes, not XY or YY
XXX: Usually fertile - Two X chromosomes pair; third does
not pair and is not transmitted
i.e. only X gametes, not XX
Thus, conditions are not passed on to progeny
genomic hybridization: microarrays
stained two different colors based on type of mutation to determine where large scale insertions and deletions occurpre
prenatal testing
associated with fetal abnormalilties not a diagnostic test just risk assessment
ultrasound, blood test for hormone levels, plasma protein levels
chorionic villi sampling
take placenta cells associated with fetus to test
less invasive
earlierin pregnancy
amniocentesis
requires needle
directly samples fetal cells in amniotic fluid to test for chromosomal abnormalities
preimplantation embryo diagnosis
in vitro fertilization
extracting eggs and fertilizing in vitro grow them in lab then extract a cell from embryo and use it to sequence genome and determine what alleles that embryo has
can screen for diseases, see which embryos to reimplant based on what alleles they have
use info to select for traits you want * controversial
eg. select for eye color
leads to many regulation, ethical, and societal questions
Lec 19: Epigenetics
epigenetics
heritable modifications in gene function not due to changes in the base sequence of DNA
DNA methylation
generally repressive
replication fork- dedicated methyl transferases that can copy pattern to newly synthesized dna
molecular mechanisms of epigenetics
1) dna methylation
2) covalent modification of histones (acetylation, methylation, etc)
3) non-covalent mods of histones (chromatin remodeling, histone variants)
4) non coding RNAs - transcriptional silencing
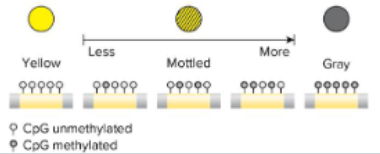
mice coat color variance due to epigenetics
agouti gene responsible for coat color
retrotransposon element insertion upstrream of agouti
IAP (cryptic promotor) can affect expression
IAP methylation → silencing of gene → brown
unmethylated IAP →yellow
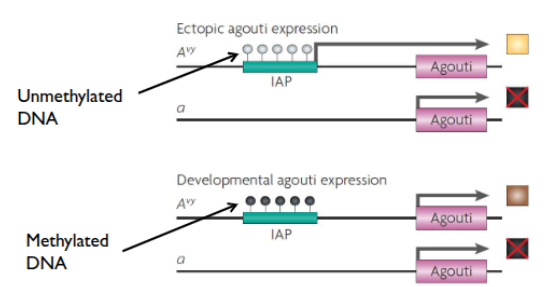
varying phenotype gradient
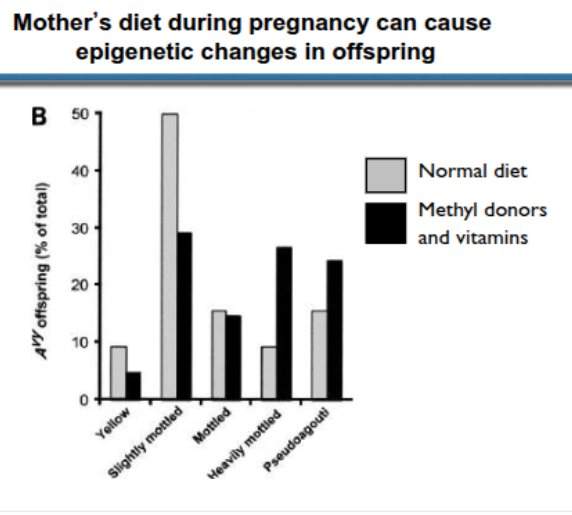
environental effects (epigenetic)
feed methyl donors, modify mothers diet → epigenetic changes in offspring
can persist in F2
epigenetics is not required for normal development. T or F?
False.
Epigenetics is required for normal development.
A very tightly controlled process of gene regulation and expression that must occur.
PRE element
with PRE: Hox gene repressed
allows cell divisions with no attrition (?) of methylation marks
no recruitment of methylation
Hox genes are still repressed
stable thru cell divisions
parental imprinting
one type of epigenetic control
occurs in placental mammals and controls development in utero
receive maternal and paternal imprints (egg and sperm)
old imprints erased during meiosis (for zygote)
new imprints
example of maternal vs paternal imprint differences
methylation on ICR region, no H19 transcription in paternal
deletions in the same part of a chromosome can lead to different effects based on paternal or maternal allele
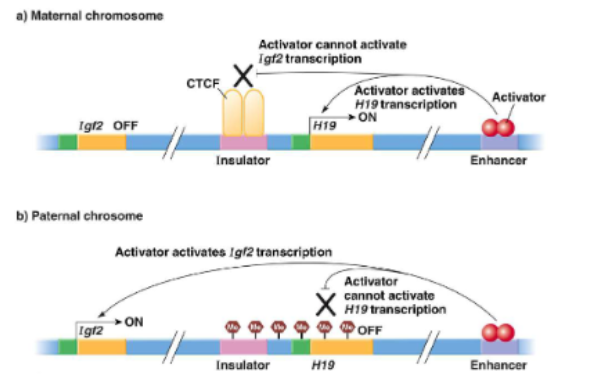
effects of mutations in imprinted genes
effect of mutation only seen in genes that are actually expressed
eg mutation in gene A is imprinted in the mom, only express the dad’s copy
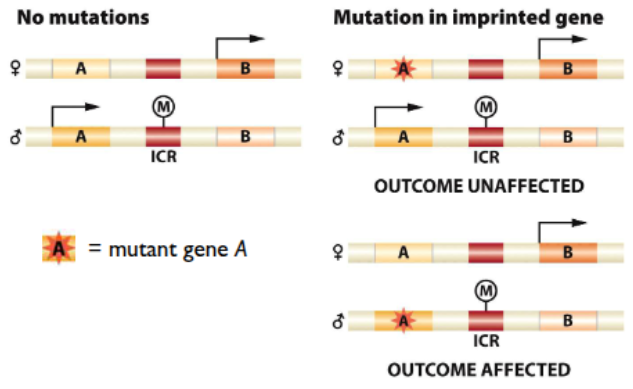
impringint of Igf2
only expressed from fathers allele
no mutation in mothers allele will have an effect
deletion inherited from dad → effect
deletion inherited from mom → no effect
Prader-Willi syndrome PWS
paternal SNRPN gene inactivated by mutation, maternal SNRPN gene inactivated by imprinting
Angelman syndrom
same part of chromosome 15
maternal copy has deletion which inactivates AS gene, paternal copy inactive via imprinting