Week 12 - Experimental Methods/Strategies in Genetics
1/37
Earn XP
Description and Tags
Chapter 14
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
38 Terms
What are model organisms and what makes each one useful for genetic studies?
Model organisms are species commonly used in labs to study genetics because they:
Have short generation times
Are easy to grow or manipulate in labs
Offer valuable insights into genetics, development, and disease
Different models suit different research purposes
E. coli (Bacteria)
Best-studied prokaryote
Generation time: ~20 mins (super fast!)
Genome: 4.64 Mb, 1 chromosome (+ plasmids), ~4262 protein-coding genes
Helped us understand DNA replication, transcription, and translation
Very easy to grow in lab settings
S. cerevisiae (Baker’s yeast)
Eukaryotic single-celled organism
Generation time: 2–3 hours
Genome: 12.2 Mb, 16 chromosomes (2n=32), ~6728 genes
Can be cultivated as easily as E.coli, but has organelles (like multicellular organisms)
Used for baking/fermentation for thousands of years
Good model for studying basic eukaryotic cell biology
C. elegans (Nematode worm)
Simple multicellular organism
Generation time: 3 days
Genome: 103 Mb, 5 chromosomes (2n=10), ~20,452 genes
~300 neurons, simple nervous system
First animal to have its whole nervous system mapped (connectome) and one of the simplest multicellular life forms
Useful for studying simple rudimentary behaviours (feeding behaviour, locomotor activity, how it responds to stress like heat shock)
D. melanogaster (Fruit fly)
Classic genetics model!
Generation time: 2 weeks
Genome: 169 Mb, 4 chromosomes (2n=8), ~14,217 genes
Helped us understand autosomal and sex-linked inheritance, genetic mapping
Many neuronal circuits characterized for behaviors like courtship, aggression, memory/learning, sensory systems and feeding
Has ~100,000–200,000 neurons

M. musculus (House mouse)
Best-studied mammal
Generation time: 10 weeks
Genome: 2731 Mb, 20 chromosomes (2n=40), ~22,322 genes
Shares many biological systems with humans
Has many tools for genetic and neurological manipulation
~71 million neurons
Great for studying mammalian genetics, disease models, and brain function

Choosing a Model Organism
Pick the simplest organism that can answer your research question
E. coli / S. cerevisiae: Best for studying basic cell processes
C. elegans: Good for studying physiological patterns of neuronal circuits for simple nervous
Drosophila (fruit fly): Complex anatomical systems + short generation time
Mouse: Closest physiology to humans, useful for complex human systems like the brain
What is forward genetics?
Forward genetics is the process of identifying genes based on mutant phenotypes.
Example: Discovery of the white gene in Drosophila
Steps:
Expose organisms to mutagens
Breed them with wild-type organisms
Screen offspring for noticeable mutations through selective breeding
What is the goal of a forward genetic screen in Drosophila?
To identify genes involved in a biological process by observing abnormal phenotypes in offspring after mutagen exposure and selective breeding.
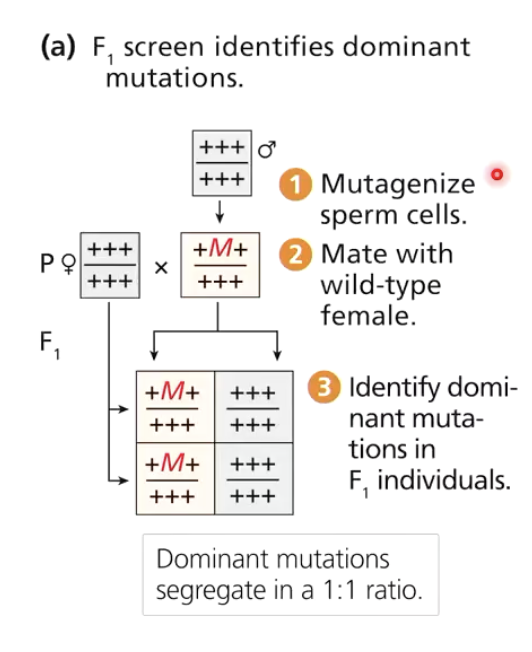
How are dominant mutations identified in forward genetics?
Mutagenize males and mate with wild-type females
Males do not produce recombinant gametes, only females undergo recombination when they produce their eggs
Easier if mutation is dominant
Dominant mutations show up in F1 generation
Take those mutants and breed together to produce F2
These are easy to detect and can be bred to form pure stocks
Dominant mutations are rare
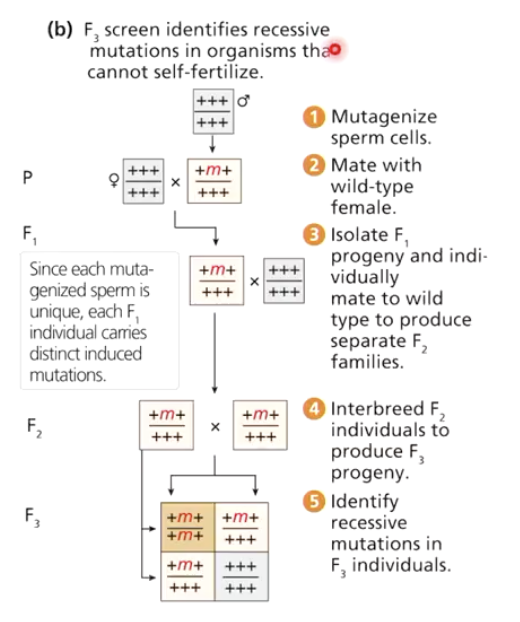
How are recessive mutations identified in forward genetics?
Mutagenize males and cross with wild-type females
F1: carry mutation heterozygously, so appear normal
Cross F1 with wild-type → F2
Interbreed a bunch of F2 to get F3
Screen F3 for homozygous recessive mutants, and use to create pure-breeding stock
Recessive mutations are more common but harder to detect
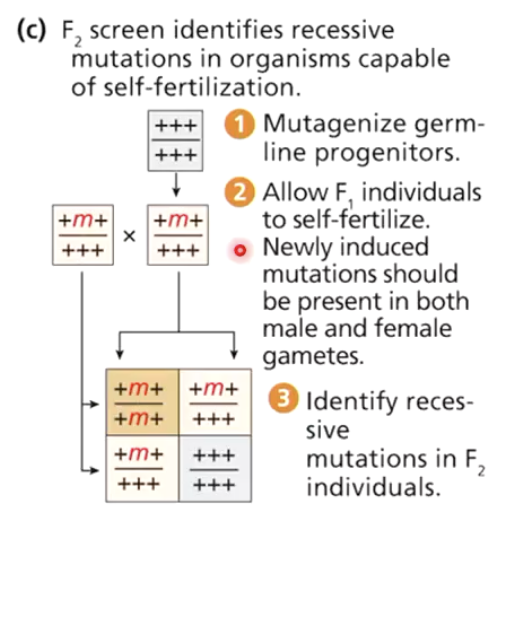
Why is identifying recessive mutations easier in self-fertilizing organisms?
Self-fertilization allows faster identification of recessive traits because you don’t need to do multiple generations of crosses
Not possible in Drosophila, but possible in organisms like yeast or C. elegans
What are Attached-X chromosomes in Drosophila, and how do they help identify X-linked mutations?
Attached-X are phenotypically females but have both X chromosomes inherited together
Useful for tracking X-linked mutations because X-linked male mutants can pass their X chromosome directly to sons (normally males give Y to sons)
Helps study X-linked inheritance patterns more efficiently
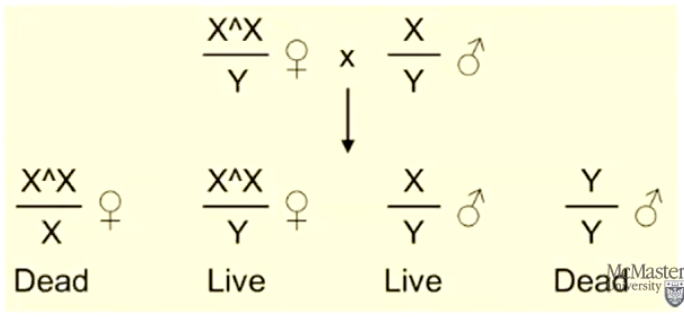
How do you determine the number of different genes mutated in a forward genetic screen?
Use a complementation test
Cross two recessive mutants:
If wild-type offspring: mutations are in different genes (2 separate genes)
If mutant offspring: mutations are in the same gene
What is reverse genetics?
Reverse genetics starts with a known gene and alters or disrupts it to study the resulting phenotype.
Opposite of forward genetics
Useful now that entire genomes are sequenced
What are common reverse genetics techniques?
Mutagenesis through gene editing (e.g., CRISPR)
Gene knockout (completely disabling the gene)
Gene knockdown (reducing gene expression)
Reporter genes (tagging a gene to study where and when it's active)
What is CRISPR-Cas9 and how does it work for gene editing?
CRISPR-Cas9 is a gene editing system where:
Cas9 is a nuclease that cuts double-stranded DNA.
It uses a guide RNA (gRNA) to find and bind to a specific DNA sequence.
Multiple gRNAs can be used to make multiple cuts in the genome.
After cutting, the cell repairs the break using:
Non-Homologous End Joining (NHEJ) — often introduces errors
Homologous Recombination (SDSA) — can copy a correct sequence from a template
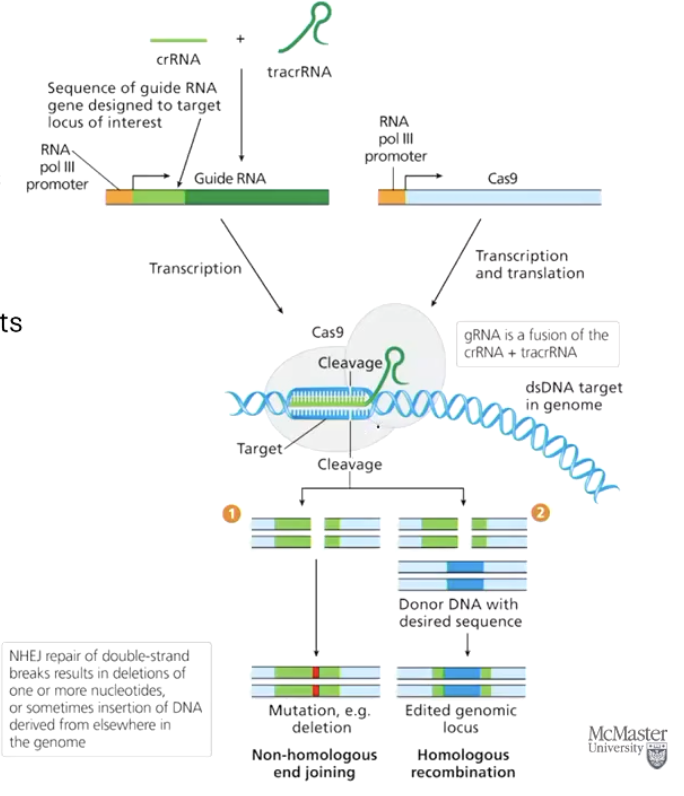
What components must be injected to perform CRISPR gene editing in an embryo?
Engineered guide RNA (to direct Cas9 to the correct DNA target)
Cas9 mRNA (so the embryo can make the Cas9 enzyme)
Donor DNA template (used for precise editing via homologous recombination)
What is a gene knockout, and how is it achieved using CRISPR?
A gene knockout means removing or disabling the coding region of a gene.
Using CRISPR:
Cas9 cuts the gene at a target site.
A template with a selectable marker (like a gene that alters phenotype) is inserted.
The marker replaces the deleted gene, allowing researchers to confirm the knockout by checking for the marker's effect.
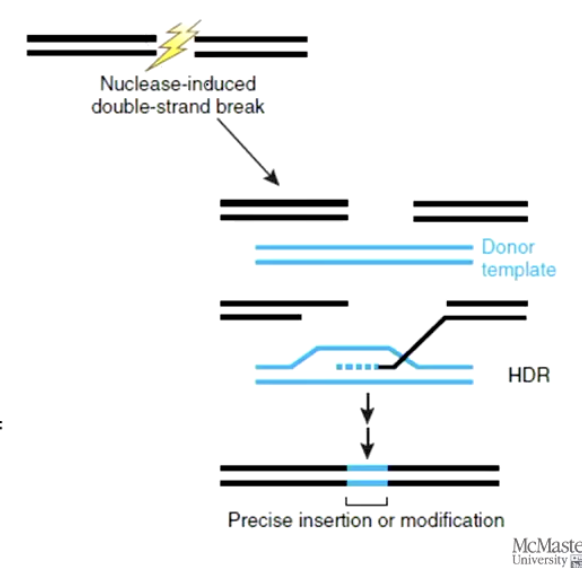
What is a gene knockdown, and how is it different from a knockout?
A gene knockdown reduces the expression of a gene (does not delete it).
Achieved using RNA interference (RNAi):
Insert a gene that makes RNA complementary to the gene of interest’s mRNA.
The matching RNA binds to the mRNA and causes it to degrade.
Less mRNA means less protein is made.
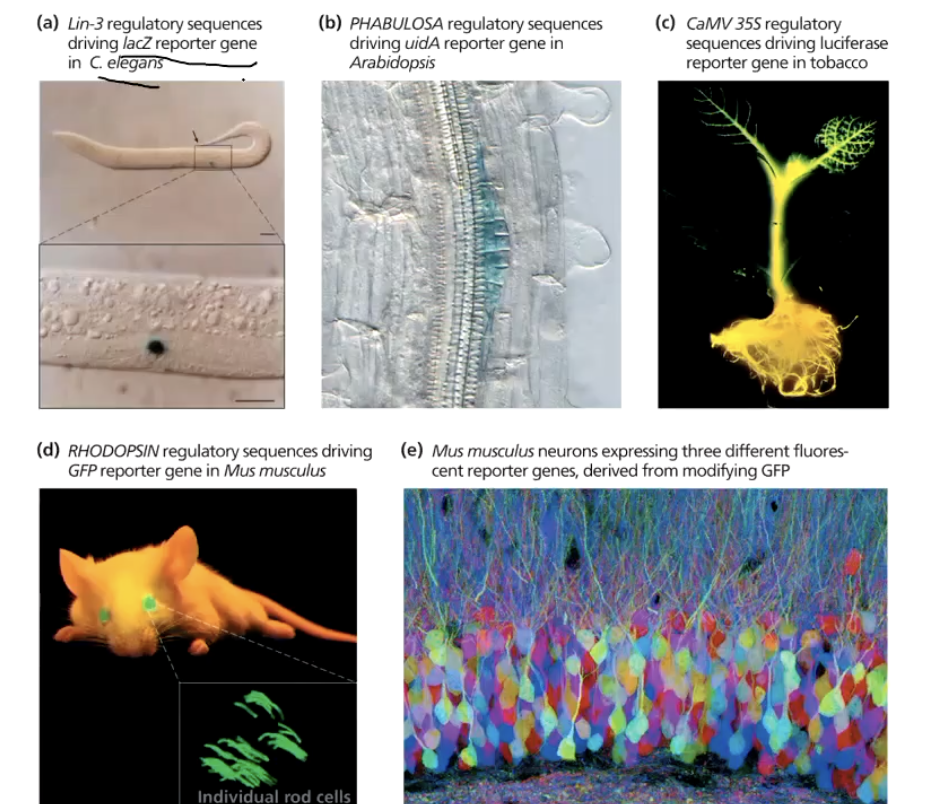
What is a reporter gene and what is it used for?
A reporter gene makes a protein that is easy to detect directly or indirectly.
It helps scientists see when and where a gene is active in an organism.
Common examples:
GFP (Green Fluorescent Protein): Glows under UV light
lacZ (beta-galactosidase): Turns blue when it breaks down X-gal (a lactose mimic)
Used in microscopic life forms
How do reporter genes help visualize gene activity?
The reporter gene is added to the DNA near or within a gene of interest.
When the gene of interest is transcribed/translated, the reporter gene is too.
You can then see the presence, location, and timing of gene activity using the reporter’s signal (e.g., GFP glowing, lacZ turning blue).
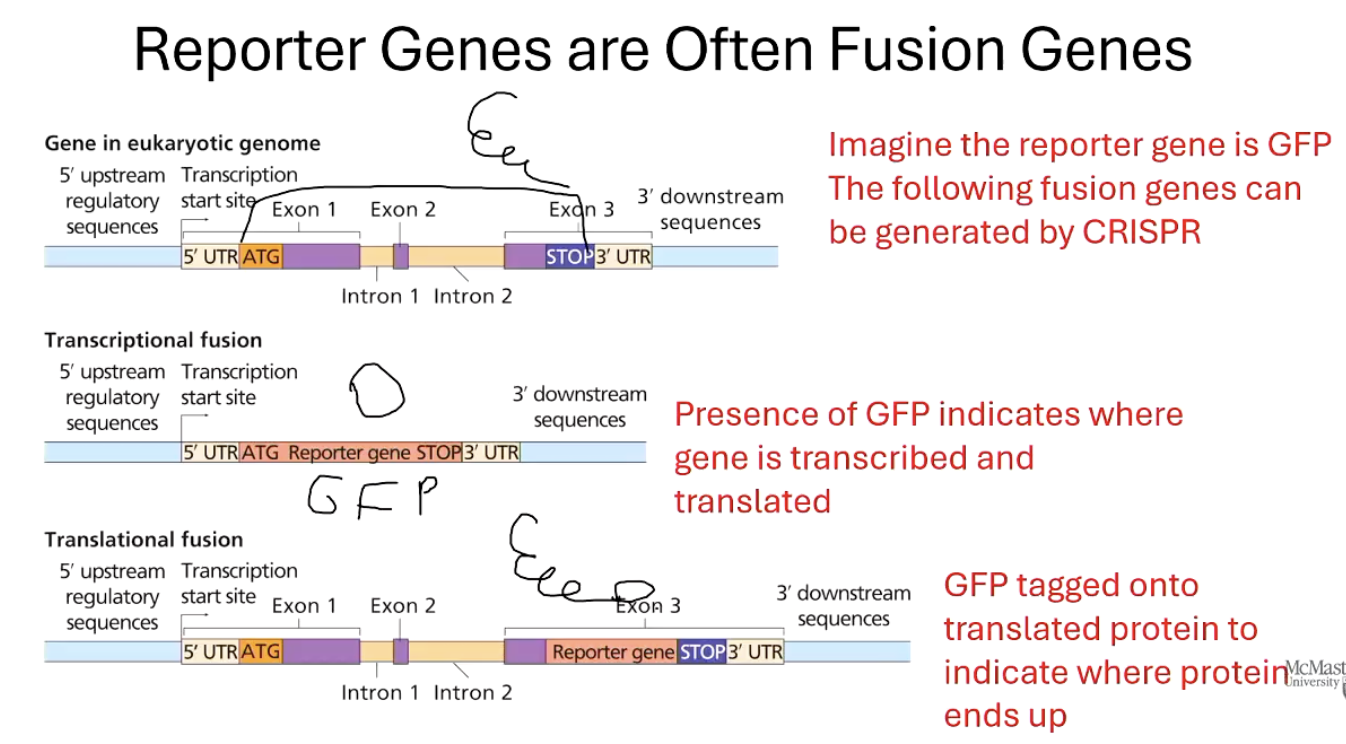
What are fusion genes and how are they used with reporter genes?
A fusion gene joins a reporter gene (like GFP) to a gene of interest.
CRISPR can be used to fuse GFP to the endogenous gene (the natural version in the genome).
When the gene is active, GFP is made together with the target protein.
This shows both where the gene is transcribed and where the protein goes in the cell.
Reporter Systems:
What is enhancer trapping and how does it work?
Enhancer trapping involves inserting a transgene into the genome with a weak promoter that cannot drive strong gene expression on its own.
If the transgene is inserted near an endogenous enhancer, the enhancer can activate the weak promoter.
This can cause high and spatially specific expression of the transgene, revealing enhancer activity in different parts of the organism.
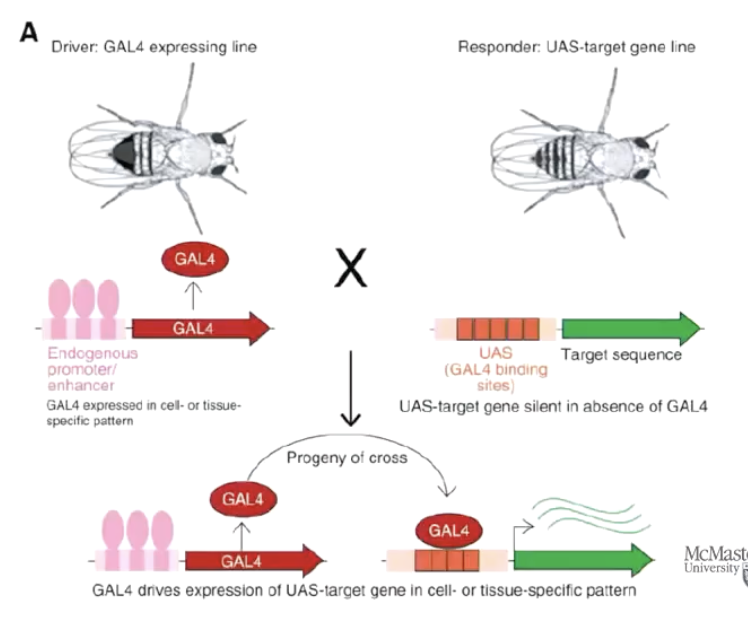
What is the GAL4-UAS system and where is it originally from?
The GAL4-UAS system is a genetic tool from yeast used to control gene expression.
GAL4 is a yeast transcription factor protein.
UAS (Upstream Activating Sequence) is a DNA sequence that GAL4 binds to.
In Drosophila (fruit flies), scientists use this system to precisely control when and where genes are turned on.
Why is the GAL4-UAS system effective in Drosophila?
Flies don’t naturally make GAL4 protein and don’t have any proteins that bind to the UAS sequence.
This means scientists can insert these components without interference.
Gene expression is only activated when both GAL4 (the driver) and UAS (the responder) are present in the same fly.
You can put any gene of interest downstream of UAS sequence, insert that into fly genome as trans genome and have GAL 4 express under the regulation of any promotor.
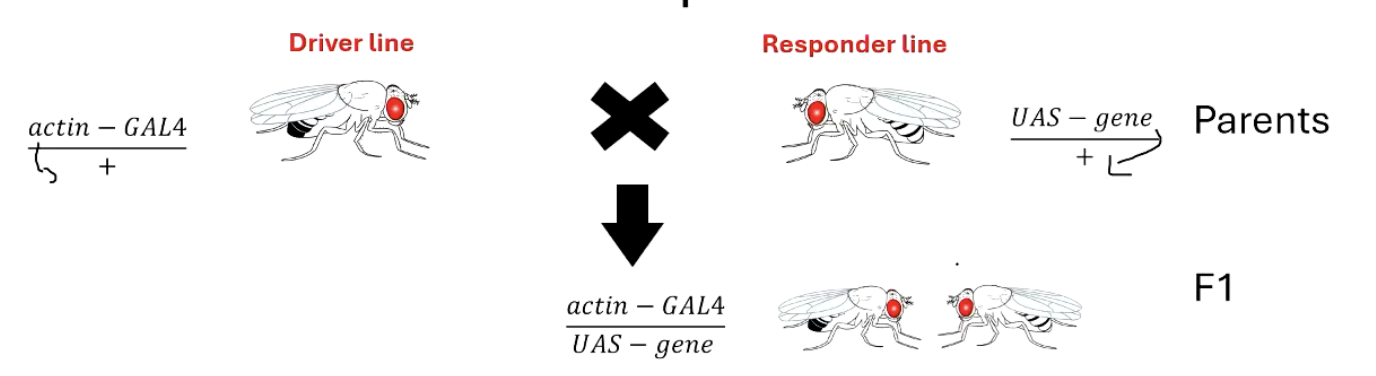
What are the two main components of the GAL4-UAS system and how do they work together?
Thousands of transgenic fly lines have either:
Driver line: A fly genetically modified to express GAL4 under the control of a specific promoter (e.g., for a brain region or tissue). - homozygous
Responder line: A fly with a target gene placed downstream of a UAS sequence. - homozygous
On their own, these flies don’t show gene expression. But when crossed together, the offspring have GAL4 binding UAS, activating the target gene in specific tissues.
Why is the GAL4-UAS system considered the “Swiss Army Knife” of behavioral genetics?
There are thousands of GAL4 driver lines targeting different tissues, neurons or specific localized cells.
There are also thousands of responder lines for different functions like reporting, manipulate gene expression or manipulate neurons.
These lines can be mixed and matched, and custom lines can be created using CRISPR.
This makes the system incredibly flexible for genetic manipulation in Drosophila.
What are some common applications of the GAL4-UAS system?
Reporter system: Use GAL4 to express GFP or lacZ to visualize where genes are active.
Gene knockdown: Use GAL4 to activate RNA interference (RNAi) to silence specific genes.
Over-expression: Use GAL4 to express endogenous gene at higher amounts.
Rescue experiments: Use GAL4 to express a gene in knockout mutants to reintroduce a normal gene to see if normal function returns.
Neuronal manipulation: Use GAL4 to express proteins that activate or silence neurons, in specific tissues.
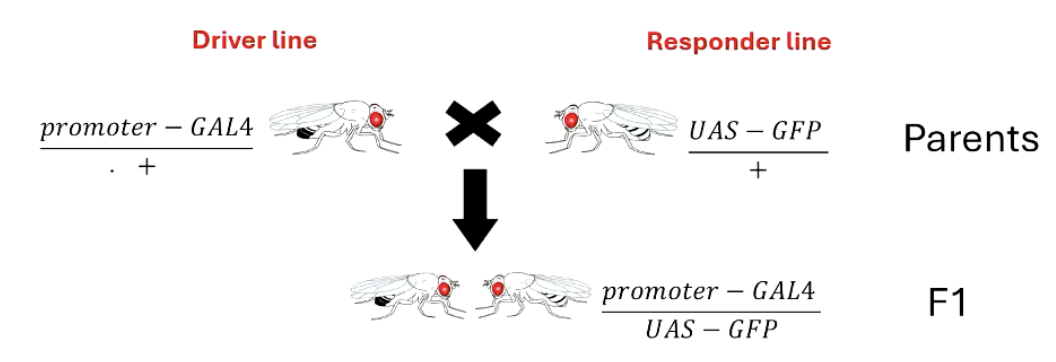
How is the GAL4-UAS system used as a reporter system?
A promoter-GAL4 driver line is crossed with a UAS-GFP responder line.
If the promoter is specific (e.g., neuron-specific), GFP is only expressed in those neurons.
If the promoter is general (e.g., actin), GFP is expressed in every cell.
This allows researchers to visualize where a gene of interest is active.
**generally driver and responder lines are homozygous, but can be heterozygous
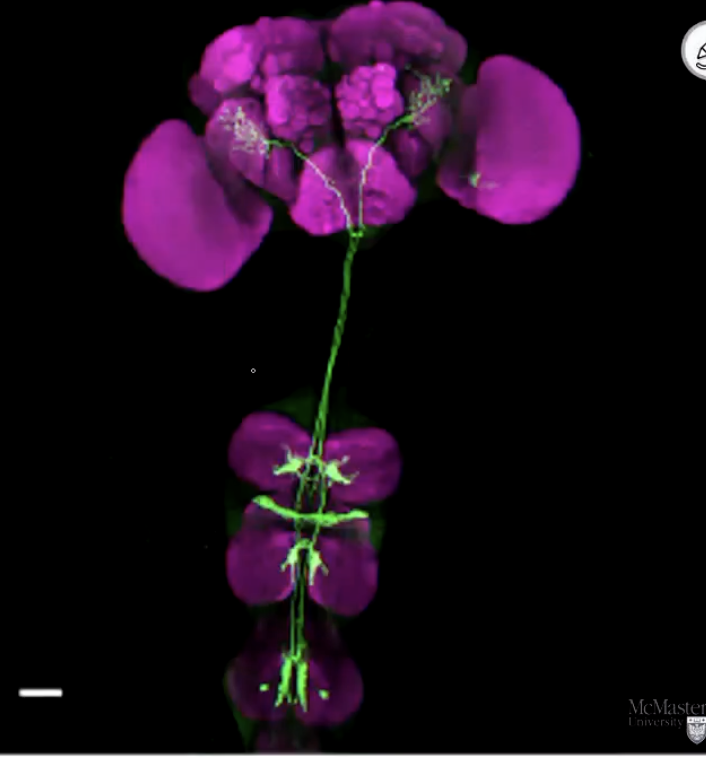
Not testable? Example of GAL4 Reporter
GFP fluorescence tagging a specific population of neurons projecting from the ventral nerve cord to the brain in Drosophila
Result of one GAL4 driver
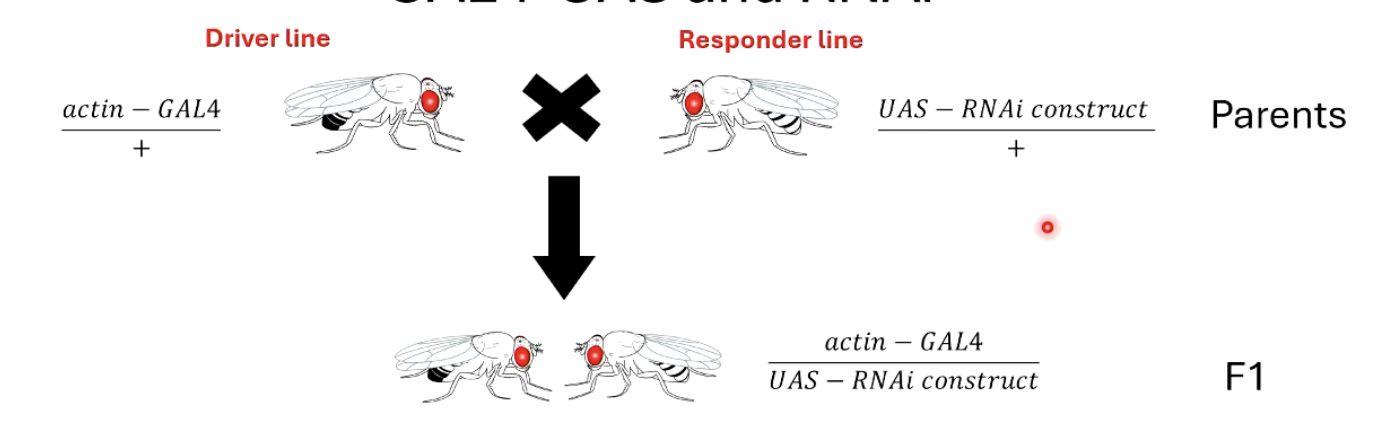
How is RNA interference (RNAi) controlled using the GAL4-UAS system?
Cross an actin-GAL4 driver line with a UAS-RNAi responder line.
The GAL4 protein activates expression of the RNAi construct in all cells.
The RNAi binds to specific mRNA, preventing translation of proteins and silencing the gene.
Using a specific promoter instead of actin can restrict knockdown to certain tissues.
Another example: inhibit pheromone production in oenocytes
How does the GAL4-UAS system allow for gene overexpression?
Cross a driver line (e.g., actin-GAL4) with a responder line containing UAS-gene.
Offspring will overexpress the gene in all cells, in addition to its natural expression (doubles gene expression)
Use of a tissue-specific promoter allows overexpression in select tissues (e.g., neurons or muscle).
Another example: upregulate pheromone genes in oenocytes
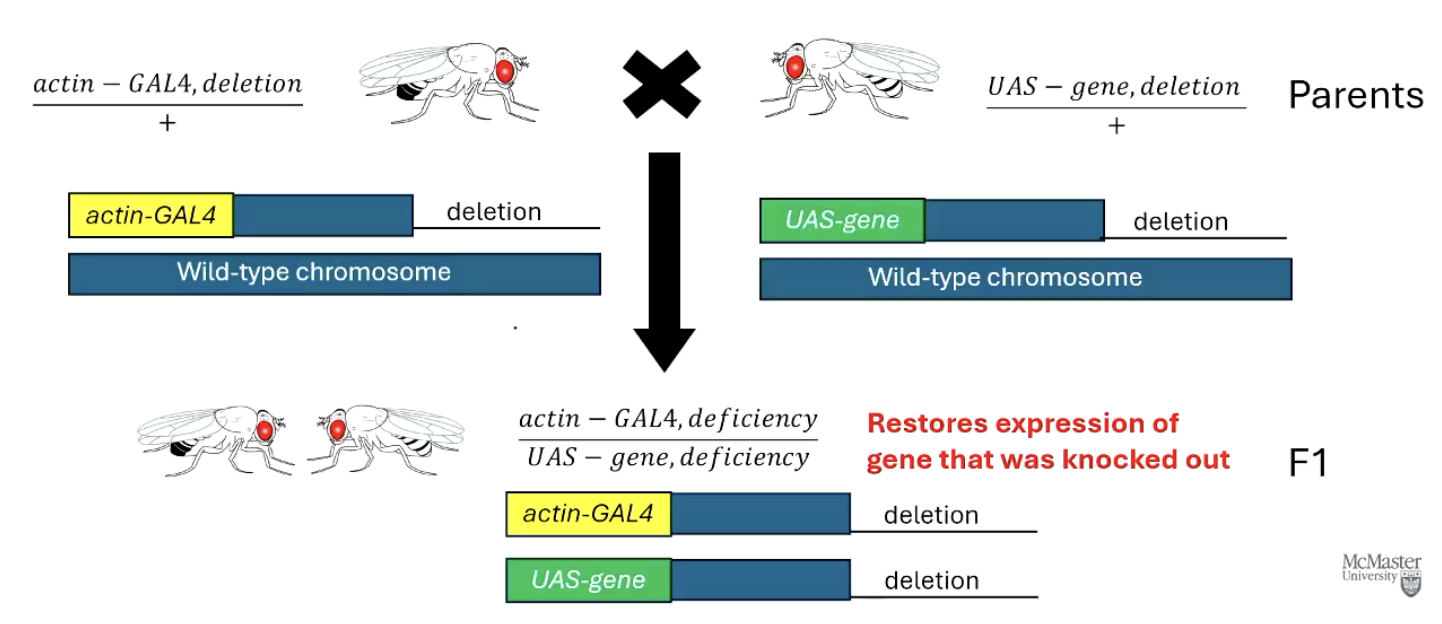
How can the GAL4-UAS system be used to rescue a gene knockout?
Use flies with a deletion (knockout) of a gene. Actin-GAL4, deletion driver and UAS-gene, deletion responder.
The UAS-gene construct restores expression of the missing gene.
If this restores normal function, it shows the gene is responsible for the lost phenotype.
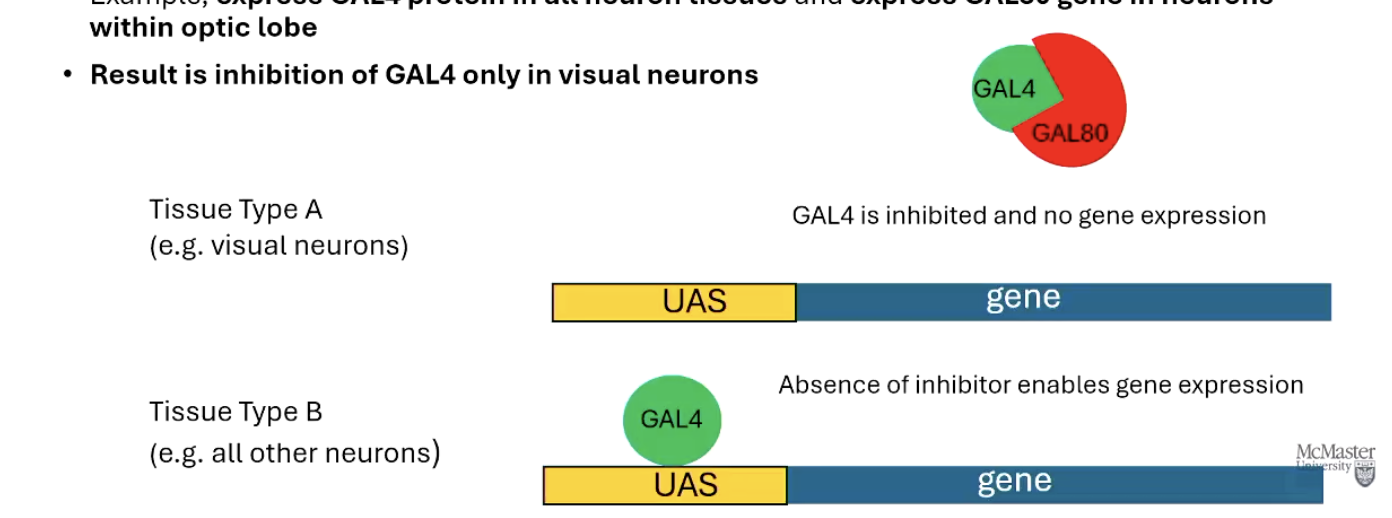
What is GAL80 and how is it used in the GAL4-UAS system?
GAL80 is a natural inhibitor of GAL4.
It can be used as separate driver to block GAL4 activity in specific tissues.
Example: Express GAL4 in all neurons, but use GAL80 in optic lobe neurons to block expression only there.
Result: Gene is not expressed in optic lobe neurons, but is in other neurons.
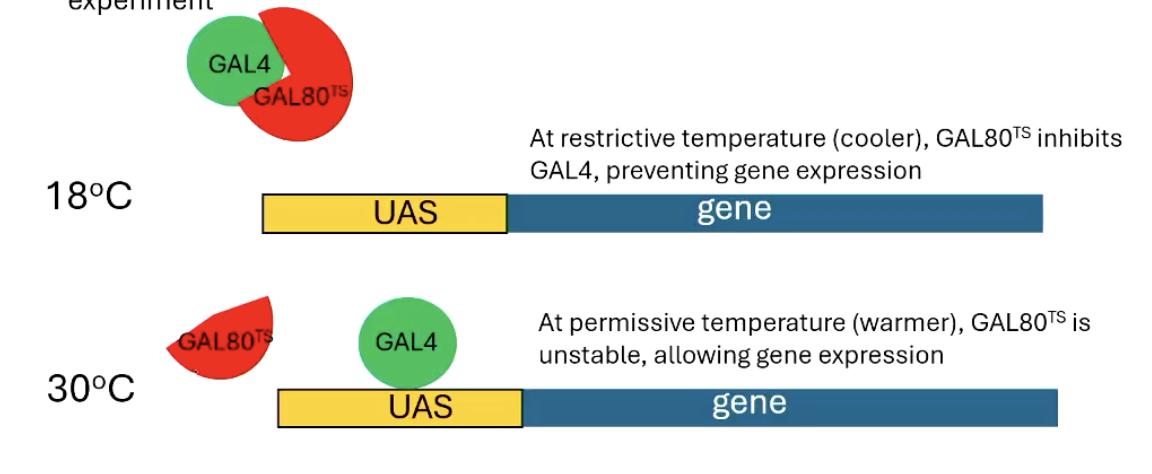
What is GAL80^TS and how does it enable temperature control of gene expression?
GAL80^TS is a temperature-sensitive version of GAL80.
At cooler temperatures (e.g., 18°C): GAL80^TS is active, blocks GAL4 → no gene expression.
At warmer temperatures (e.g., 30°C): GAL80^TS is unstable, GAL4 is active → gene expression starts.
Allows researchers to control the timing of gene expression (e.g., only during adulthood).
How can neurons be silenced using the GAL4-UAS system?
Use GAL4 to express tetanus toxin in specific neurons.
GAL4 driver, UAS-TNT responder.
Tetanus toxin blocks neurotransmitter release, silencing the neurons.
This helps determine the function of specific neurons in behavior.
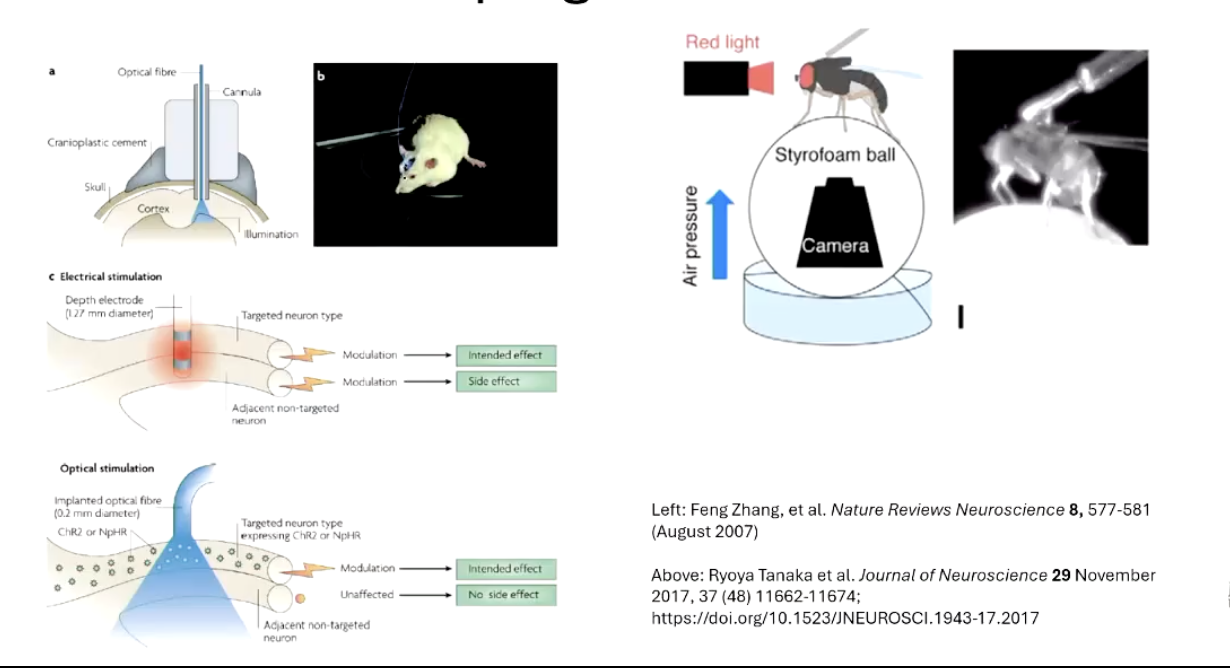
What is optogenetics and how does it work with the GAL4-UAS system?
Optogenetics involves expressing light-sensitive receptors like channelrhodopsin-2 (ChR2) in neurons.
Using GAL4-UAS, ChR2 is expressed in specific neurons.
Shining blue light on the fly activates those neurons, allowing researchers to control behavior with light.
**You do not need to know these slides for exam
Optogenetics – Sample Videos