Module 7: The Cytoskeleton
1/68
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
69 Terms
What is the cytoskeleton and what are its roles in the cell?
Network of protein-based filaments
Provides cell shape and structural support
Dynamic, not static → enables:
Cell movement
Cell growth
Cell differentiation
Composed of 3 filament types:
Actin filaments
Microtubules
Intermediate filaments
How does the cytoskeleton contribute to specialized cell structures and shape?
Crucial for maintaining cell shape, especially in differentiated cells
Examples:
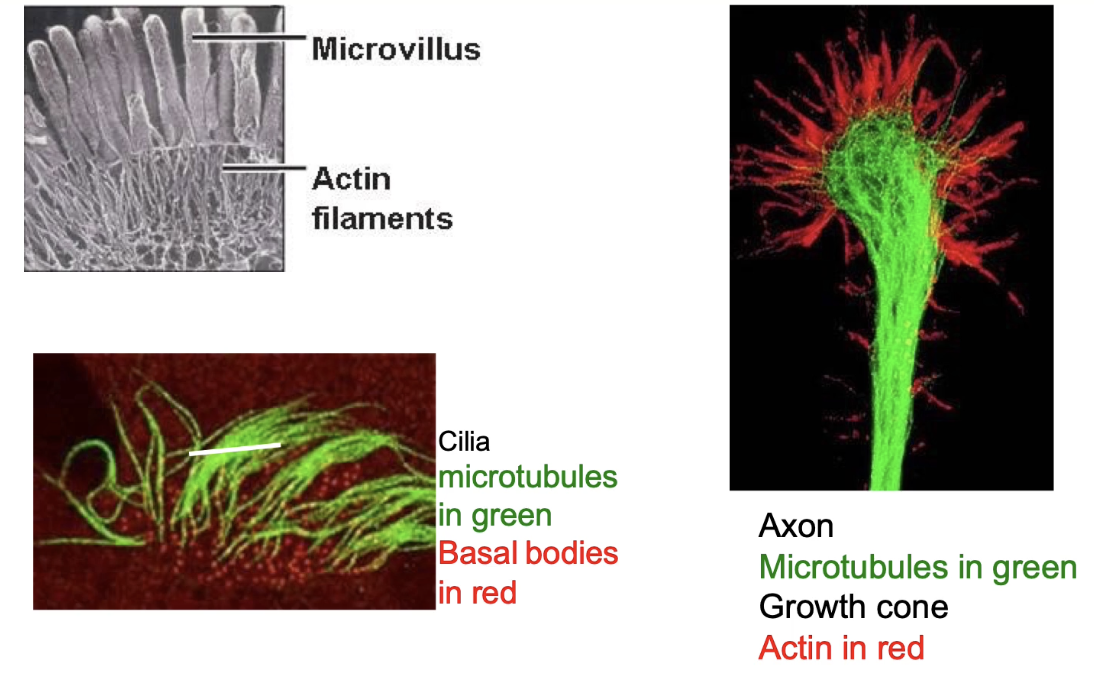
Microtubules → structure in cilia
Actin filaments → shape/function in microvilli (epithelial cells)
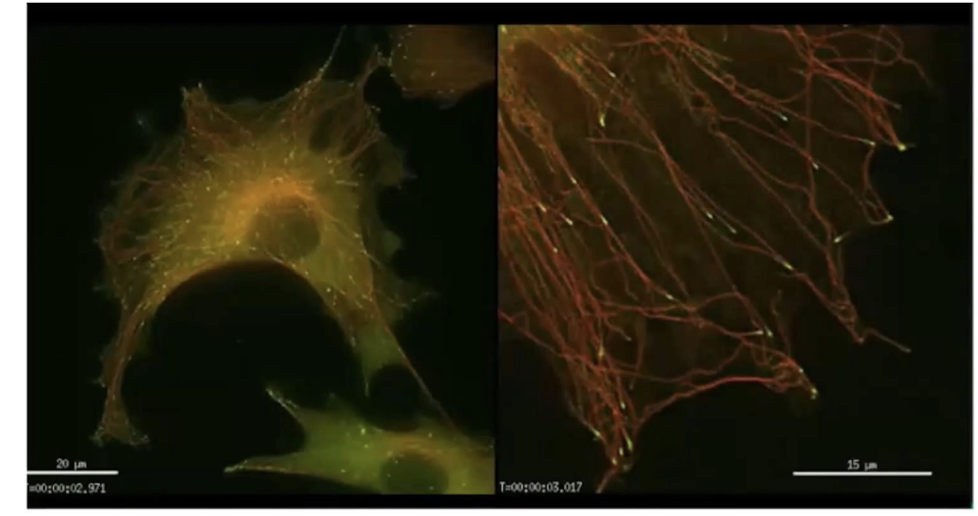
Neuron example:
Microtubules (green) → support axon
Actin (red) → shapes growth cone
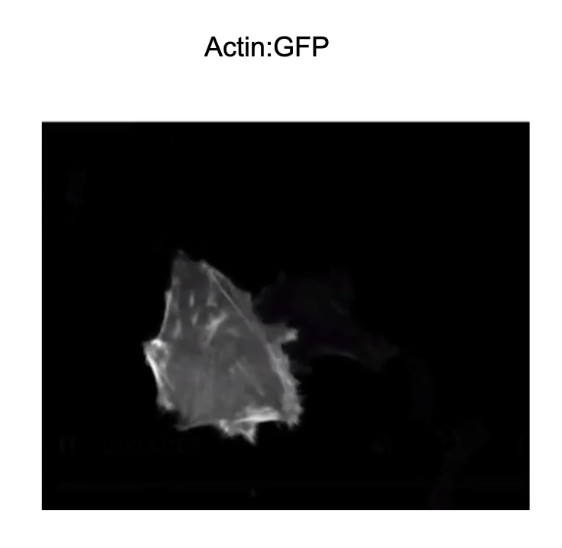
Why is the dynamic nature of the cytoskeleton important?
Essential for cell movement
Supports processes like:
Cell migration
Cell division
Example:
Ovarian cancer cell expressing actin-GFP
Shows real-time actin filament dynamics during movement
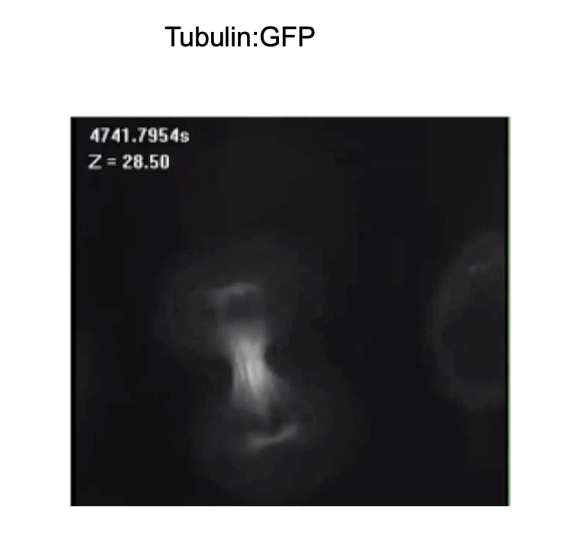
How is the dynamic nature of microtubules shown during mitosis?
Seen in cell division (mitosis)
Example:
Breast cancer cell expressing Tubulin:GFP
Visualizes microtubule spindle formation over time
Demonstrates the structural reorganization of cytoskeleton during mitosis
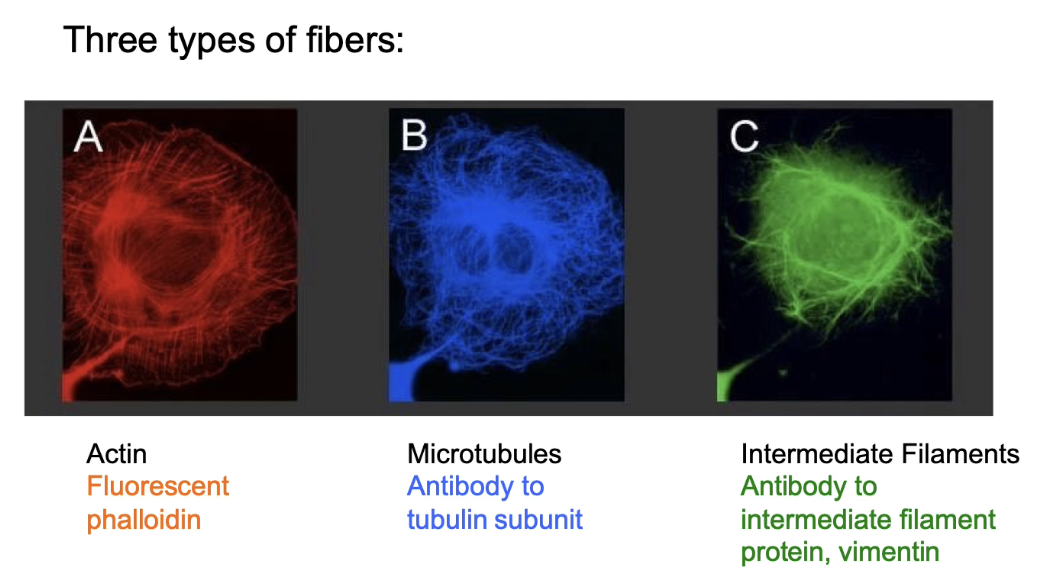
What are the 3 types of cytoskeletal fibres and how are they labeled in cells?
Types: Actin filaments (microfilaments), Microtubules, Intermediate filaments (IFs)
Defined by: Diameter and subunit type
Actin labeling:
Phalloidin (fluorescent toxin from death cap mushroom)
High specificity & affinity to actin
Stabilizes actin filaments
Antibody to actin
Actin:GFP fusion
Microtubule labeling:
Antibody to tubulin
Tubulin:GFP fusion
Intermediate filament labeling:
Antibody to filament-specific subunit
GFP fusion proteins
All 3 fibres:
Found in all eukaryotic cells
Form overlapping but distinct structures
Can be seen together in cells via fluorescent labeling
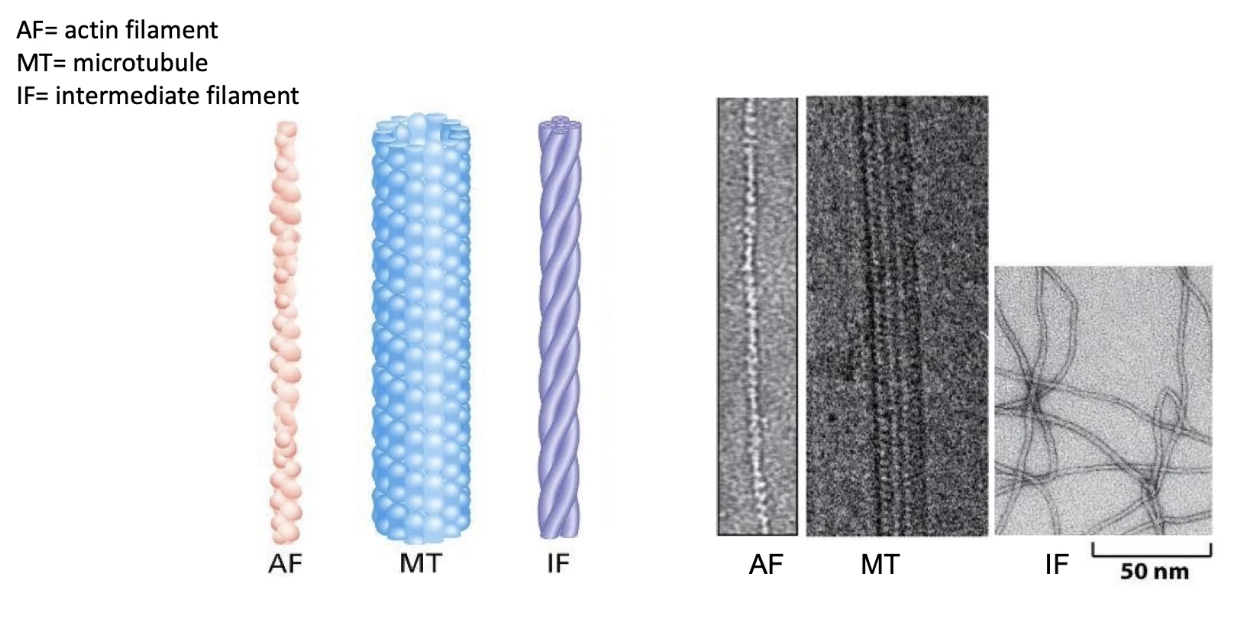
What are the structural differences among the 3 cytoskeletal fibres?
All fibres are polymers made of protein subunits
Actin filaments (microfilaments):
Thinnest
Made of monomeric actin
Microtubules:
Thickest
Made of α- and β-tubulin dimers
Intermediate filaments (IFs):
Medium thickness
Made from varied proteins, depending on cell type
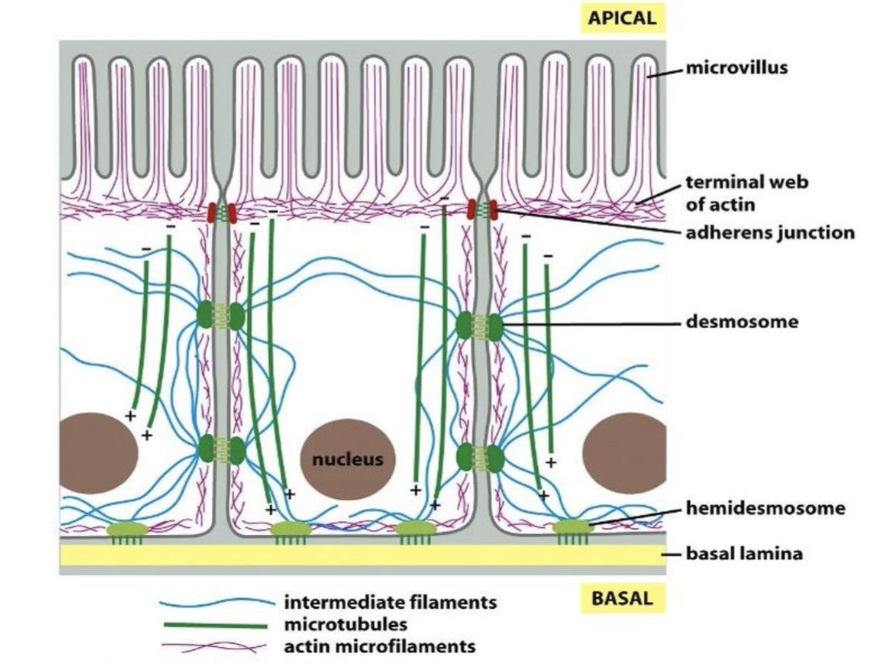
How are the 3 cytoskeletal fibres distributed in epithelial cells?
Actin (red):
Located at apical surface
Shapes microvilli
Intermediate filaments (blue):
Span the entire cell for structural support
Composed of lamin proteins
Also form nuclear lamina (supports nucleus)
Microtubules (green):
Form networks for intracellular transport
Distribution is cell-type specific, with each fibre type occupying unique regions
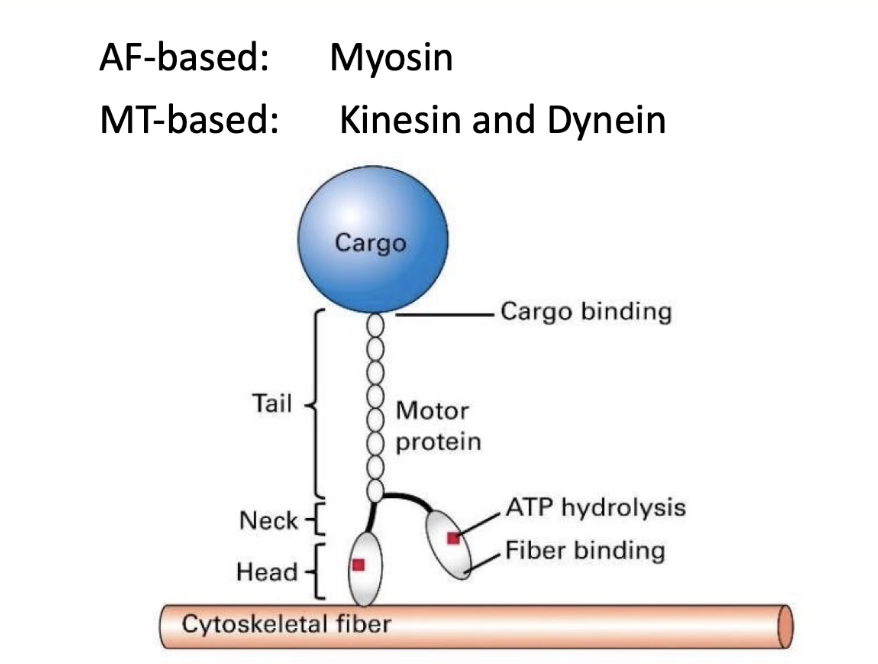
What are the motor proteins associated with cytoskeletal filaments?
Motor proteins move along actin filaments and microtubules
No motor proteins are associated with intermediate filaments
Actin-based motors:
Myosin proteins
Microtubule-based motors:
Kinesin
Dynein
General motor protein structure:
Head domain binds to cytoskeletal filament
Tail domain binds to cargo
Movement powered by:
ATP hydrolysis → drives protein “walking” or stepping
What are common actin-based cell movements and structures?
Actin density is highest at cell periphery
Functions of actin filaments:
Microvilli formation
Contractile bundles in muscle (sarcomeres)
Filopodia & lamellipodia for cell migration
Contractile ring in cytokinesis
Actin structural organization in cells:
Contractile stress fibers – throughout the cell
Gel-like network – at the cell cortex
Tight parallel bundles – in filopodia
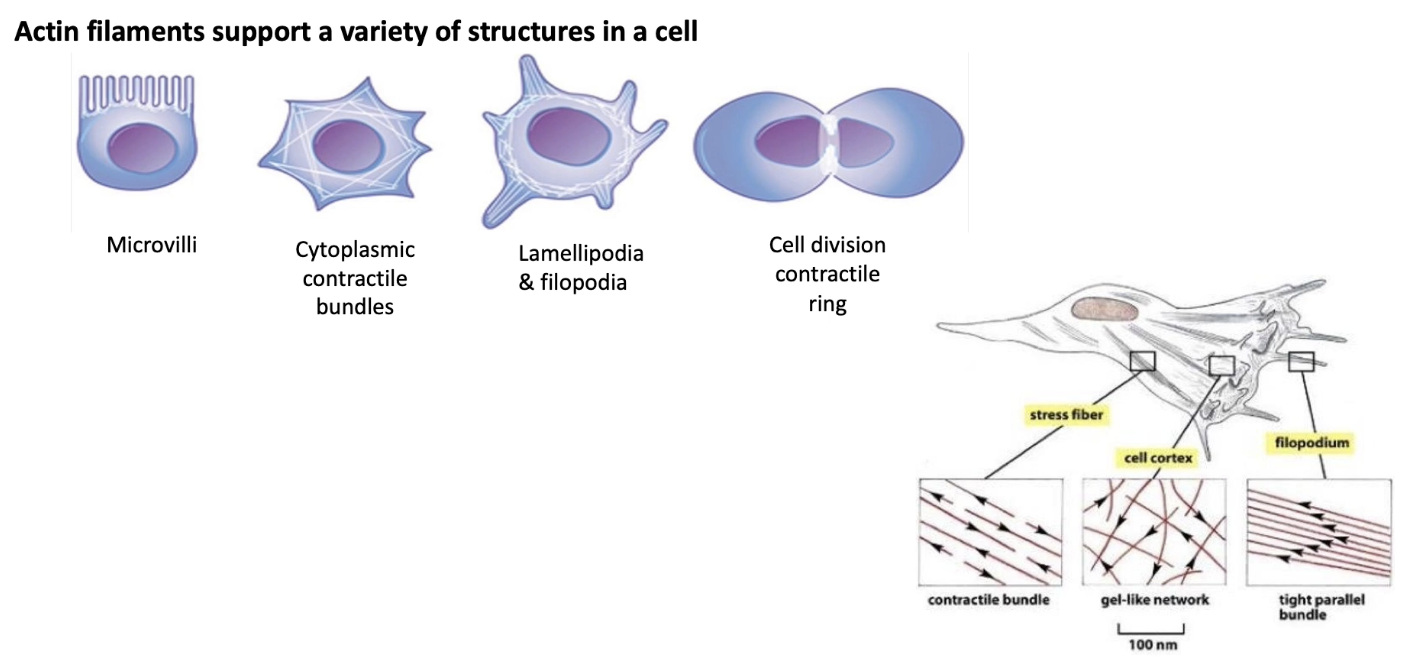
What is F-actin and what is its structure?
F-actin = filamentous actin
Made of:
Two helical strands of polymers
Each strand is a chain of G-actin (globular actin) monomers
Diameter: 5–9 nm
Structure features:
Strands spiral around each other
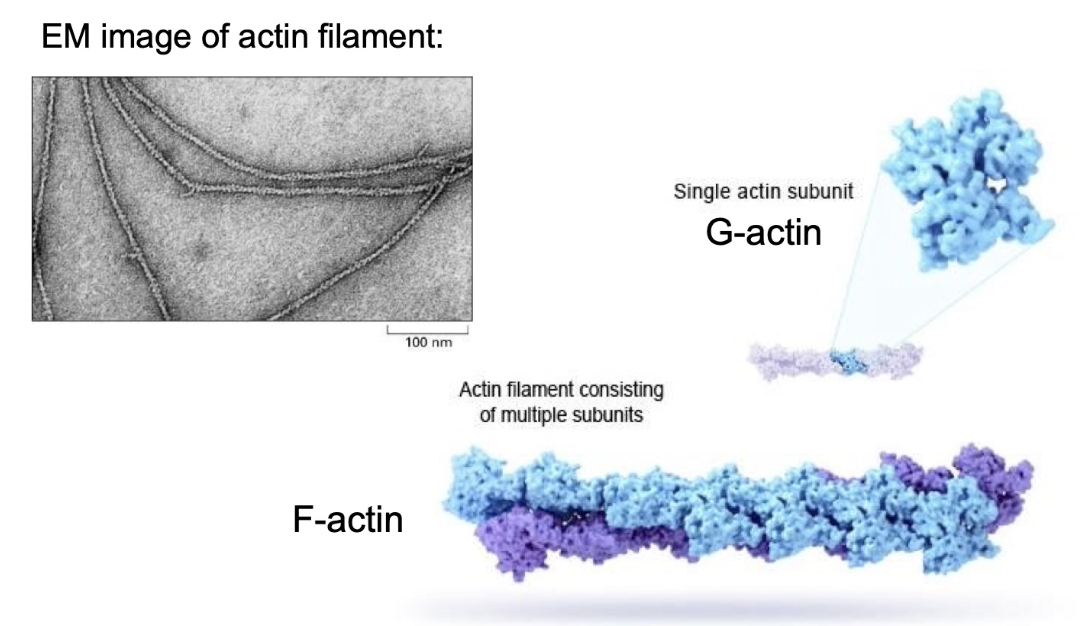
What does it mean that actin filaments are polar, and how is this polarity visualized?
Actin filaments are polar → each end behaves differently
Polarity visualized using myosin head decoration in electron microscopy
Myosin binds in one direction, showing filament orientation
Plus-end (barbed end):
Grows faster
Higher actin subunit addition
Minus-end (pointed end):
Grows slower or may shrink
Polymerization is more active at the plus-end
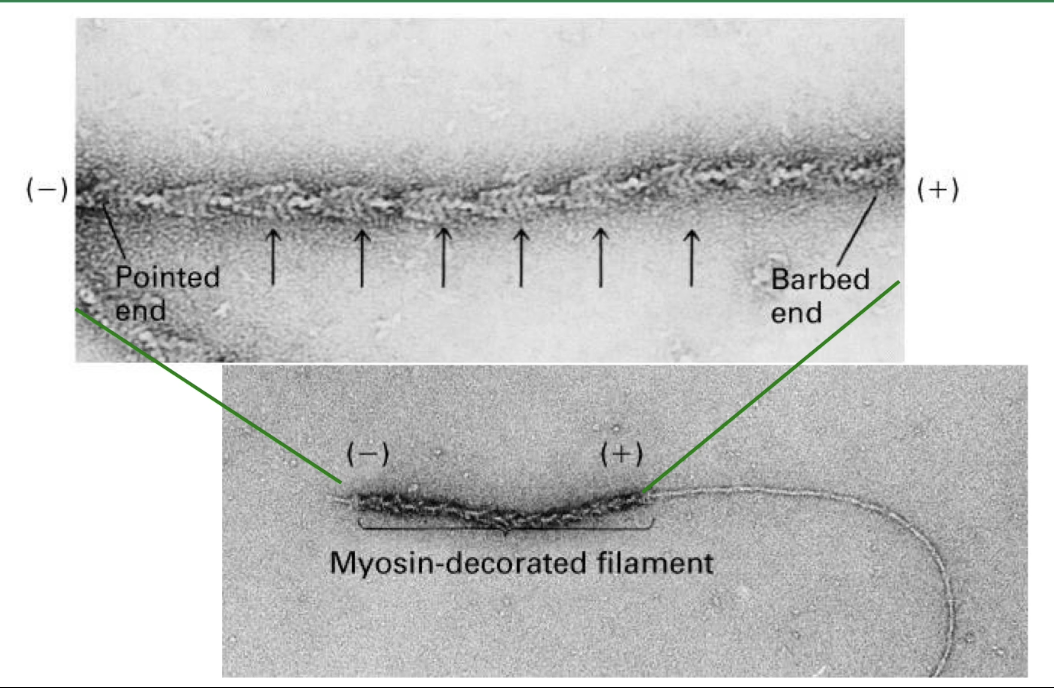
What is G-actin, and how does it contribute to actin filament polarity?
G-actin = globular actin monomer
Has 4 structural domains
ATP-binding site located in cleft between domains 2 & 4
Each monomer is polar
Filament polarity arises from monomer orientation
ATP-binding pockets face the minus end.
Inside the filament, these pockets are hidden, except at the very end
Only the last few monomers at the minus end have their ATP-binding sites partially exposed.
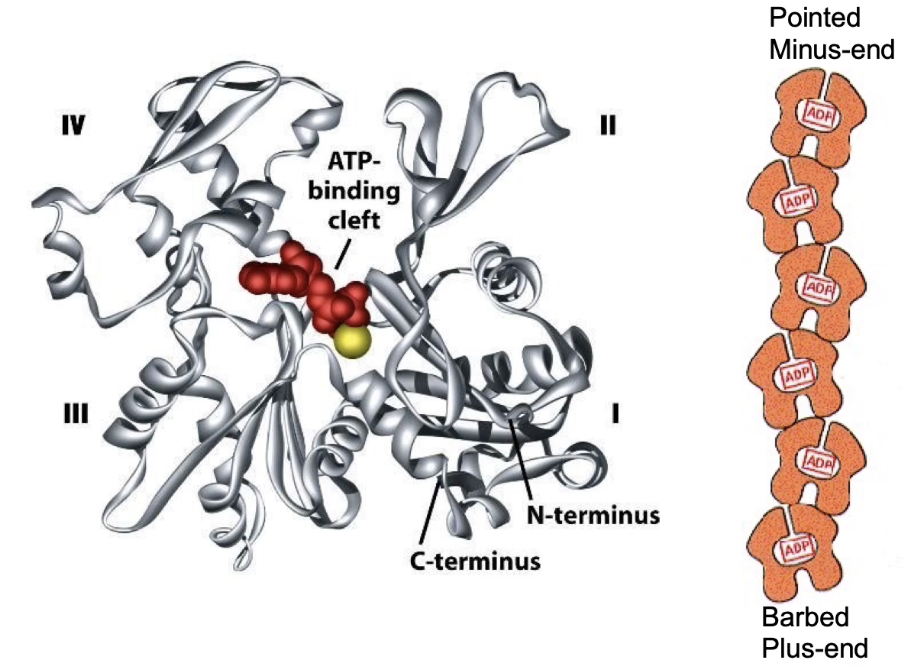
How does actin filament polymerization work and what role does ATP play?
F-actin = filamentous actin (polymerized G-actin)
Actin filaments are dynamic
Undergo both polymerization and depolymerization
Plus-end:
More growth (polymerization > depolymerization)
Minus-end:
More shrinkage (depolymerization > polymerization)
ATP-bound actin:
Adds to the plus-end
Actin has ATPase activity → hydrolyzes ATP to ADP + Pi
Most filament = actin-ADP
ADP is trapped inside filament (not released)
In cytosol:
Free actin-ADP → releases ADP → recharges with ATP
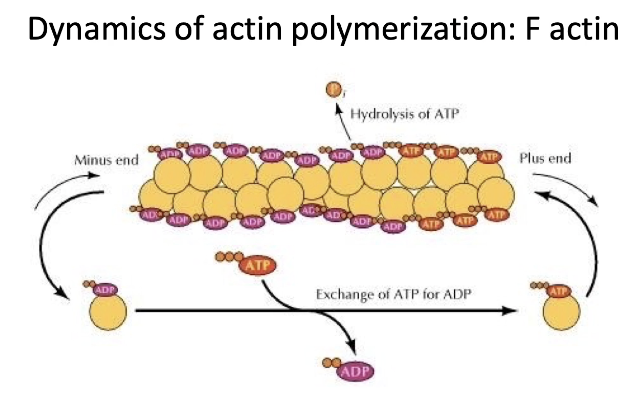
What is critical concentration in actin dynamics, and what proteins regulate actin polymerization?
Critical concentration (Cc):
Point where polymerization = depolymerization
[Actin] > Cc → filament grows
[Actin] < Cc → filament shrinks
Cc differs at plus- & minus-ends → different filament dynamics
Regulatory proteins:
Profilin:
Binds actin-ATP
Promotes ATP binding
Activates monomer → accumulates at plus-end
Thymosin:
Binds actin monomers → inhibits polymerization
Thymosin-actin dimers accumulate at plus-end and creates storage buffer of actin monomers
Capping proteins:
Block ends of actin filaments→ inhibit growth or shrinkage
What is actin treadmilling, and why is it important?
Treadmilling
Polymerization at plus-end = depolymerization at minus-end
No net length change, but filament moves forward
Important for:
Cell movement
Cytoskeletal remodeling

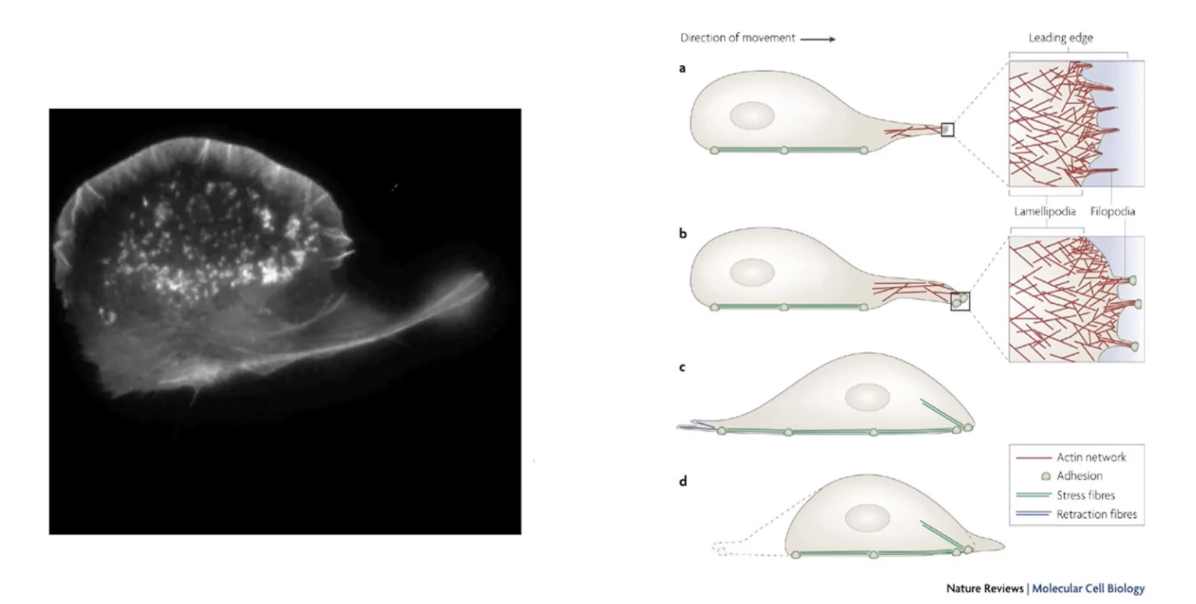
How does actin filament reorganization enable cell movement?
Reorganization of actin filaments pushes cell membrane outward
Forms two structures:
Lamellipodia = broad, fan-like membrane expansions
Filopodia = thin, finger-like projections
Steps in movement:
Leading edge forms
Lamellipodia spread (fan like expansions)
Filopodia extend
Cell moves in direction of leading edge
All require dynamic remodeling of actin
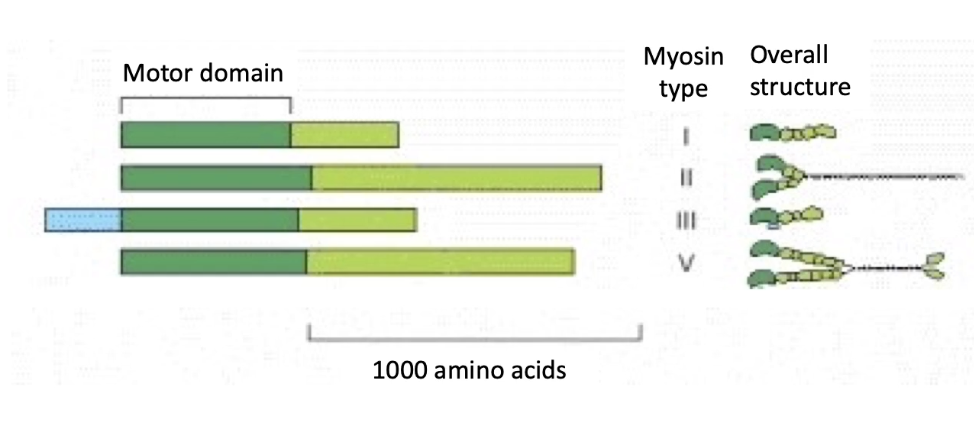
What are myosin motor proteins, and how do they interact with actin filaments?
Myosins = actin-based motor proteins
Use ATP hydrolysis to "walk" along actin filaments
Most move toward plus-end
(3/8) Key families:
Myosin I, II, V → found in most eukaryotic cells
Structure:
Head (motor) domain at N-terminus:
Binds actin
Hydrolyzes ATP
Tail domain:
Varies to carry different cargo or interact with different targets at different rates
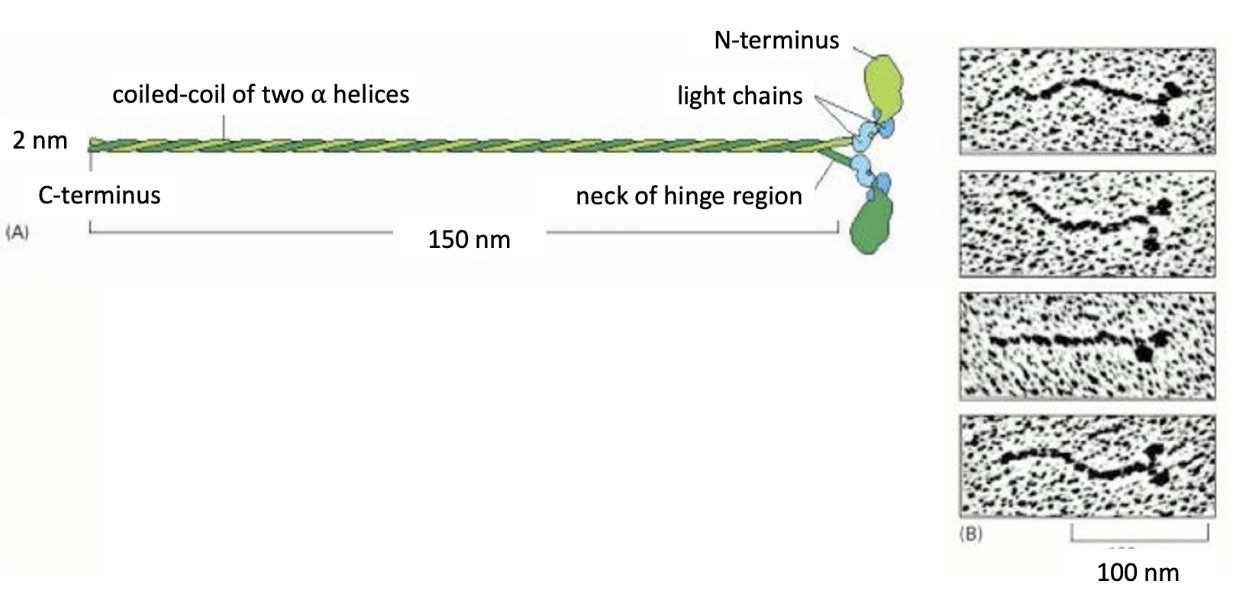
What is the structure of Myosin II, and what is its role?
Myosin II = muscle contraction motor
Structure:
2 heavy chains → form coiled-coil tail
4 light chains (2 types) → regulate activity
Motor domain (head):
Seen in EM images as globular heads
Located at top of tails
Responsible for movement along actin
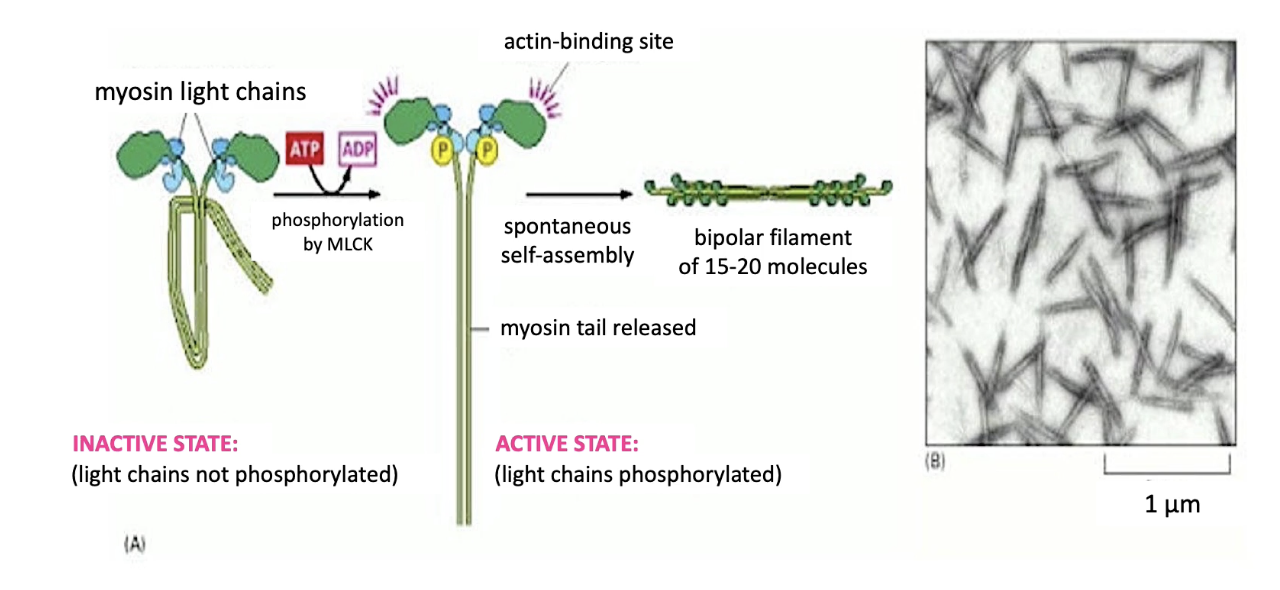
How is Myosin II activated and what is its function in cells?
Activation:
Myosin Light Chain Kinase (MLCK) phosphorylates myosin light chains
Triggers tail extension & activates actin-binding domains on motor heads
Assembly:
15–20 myosin II proteins form a bipolar thick filament
Function:
Myosin II doesn't carry cargo
Generates contractile forces essential for many cell processes
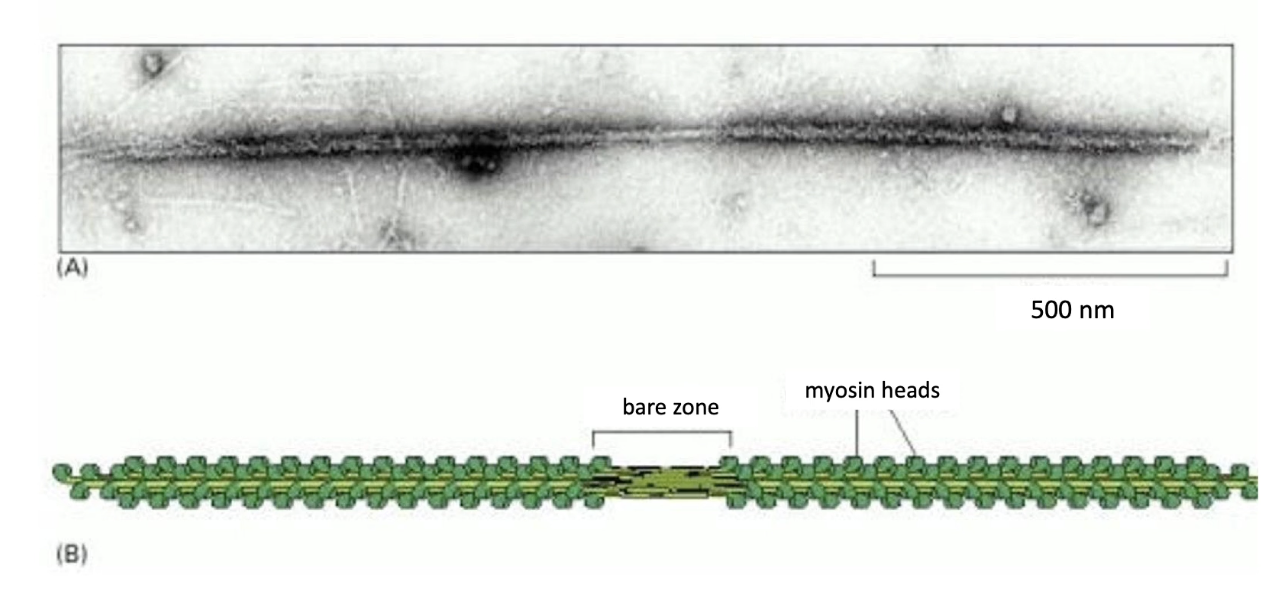
What is the structure of a Myosin II thick filament?
Bipolar filament:
Motor heads on both ends
Bare zone in the center (no motor heads)
Function of structure:
Allows motor heads to interact with actin on both sides
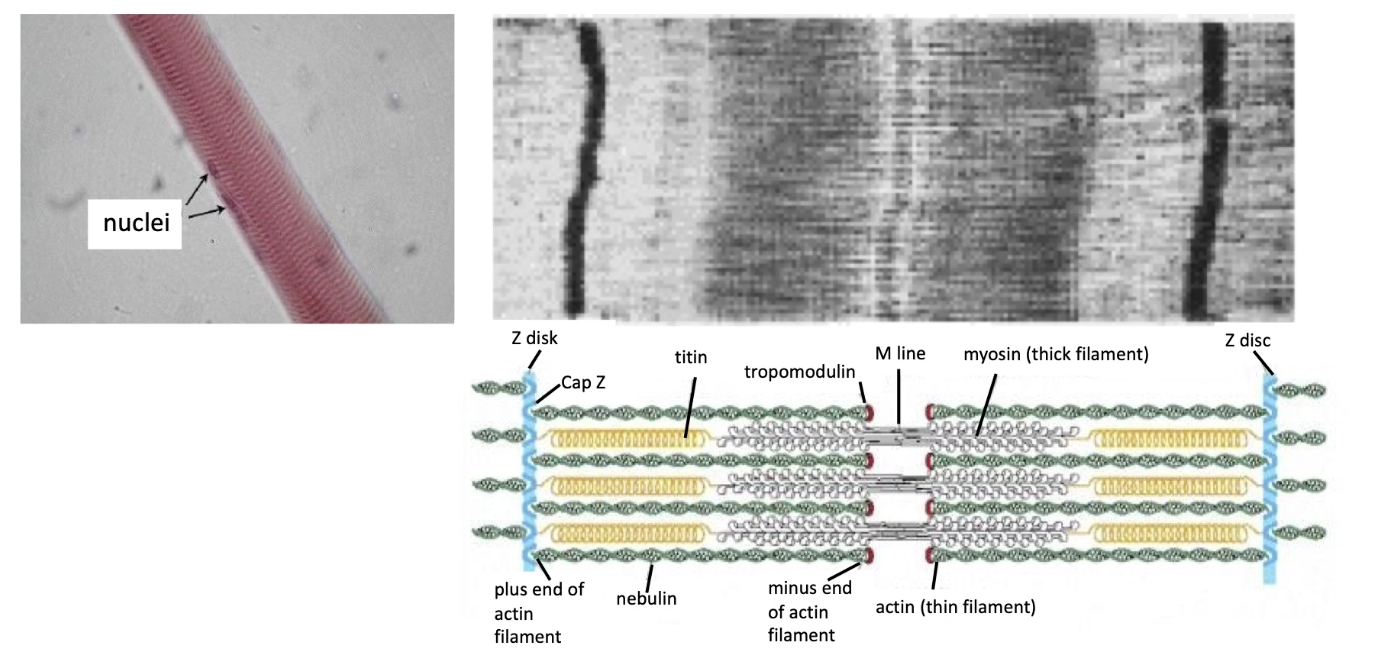
What role does Myosin II play in skeletal muscle contraction and sarcomere structure?
Myosin II + actin = sarcomere (basic unit of striated muscle)
Sarcomere components:
Z-discs anchor plus-end of actin filaments
CapZ (plus-end capping), Tropomodulin (minus-end capping)
Nebulin: stabilizes parallel actin filaments
Between parallel actin fibers there are myosin thick filaments
Titin: spring-like protein anchors myosin to Z-discs
Muscle contraction:
Myosin interacts with actin → Z-lines pulled closer together
“Striated muscle”:
Refers to striped appearance of sarcomeres under microscope
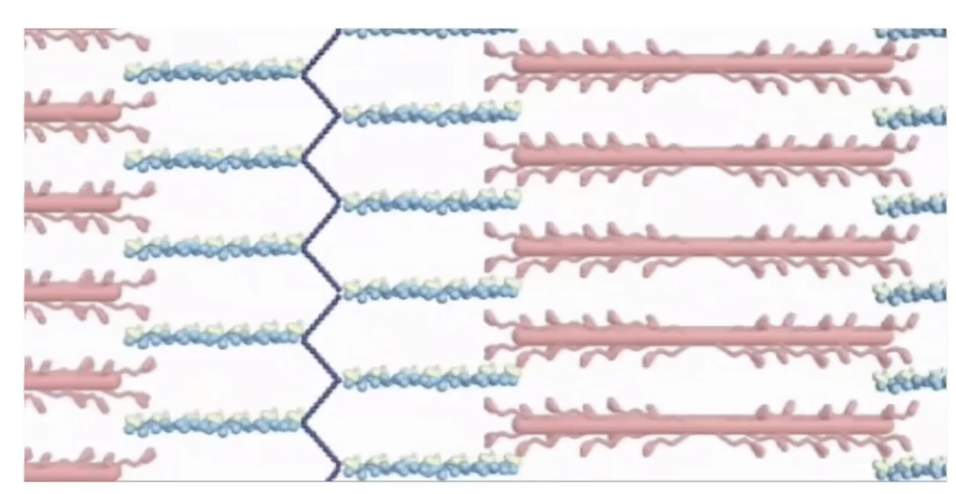
How does muscle contraction occur at the sarcomere level?
Myosin heads pull actin toward the center of the sarcomere
Due to cyclical association of actin with motor heads
Shortening of sarcomere but no change in filament length, just sliding past each other
Each myosin head:
Binds ATP → hydrolyzes it to ADP
"Walks" toward plus-end of actin
Shortening of sarcomere = muscle contraction
Calcium-dependent process:
Ca²⁺ allows myosin binding sites on actin to be exposed
Relaxation:
Ca²⁺ is removed
Myosin heads along thick filament release actin thin filaments
Thick and thin slide past and sarcomere elongates → muscle relaxes
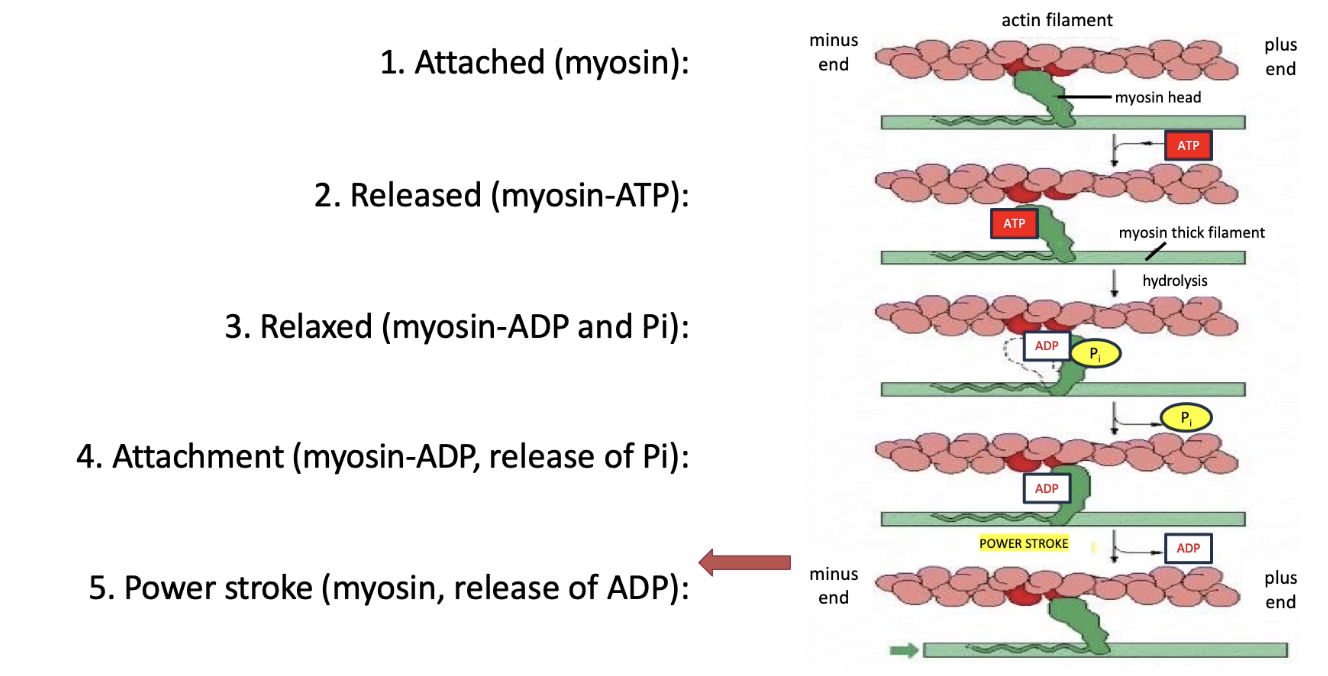
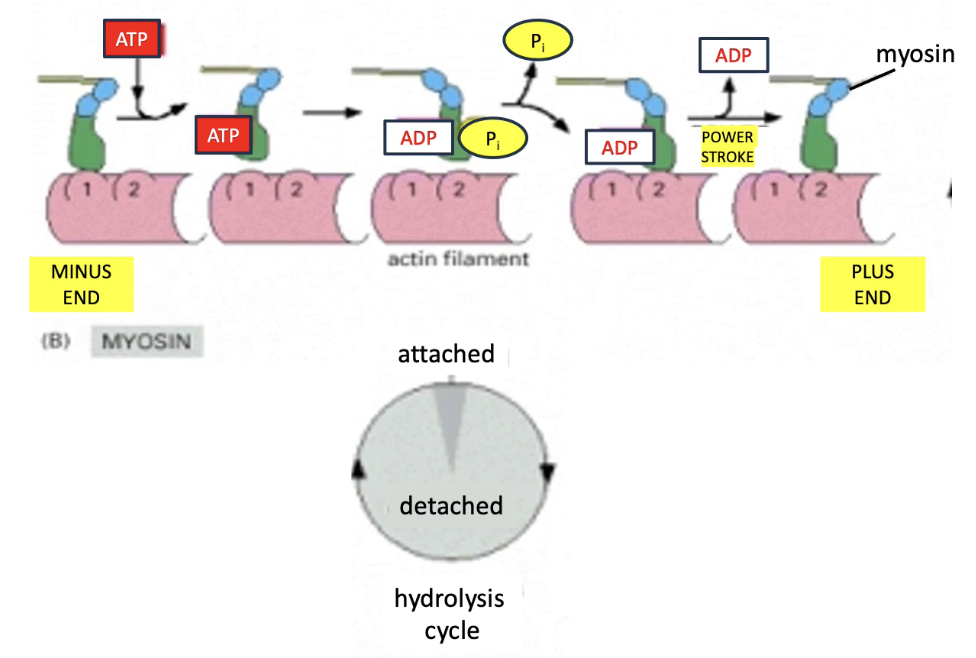
What are the steps in the myosin ATPase cycle that power muscle contraction?
Chemical energy (ATP) → Mechanical movement (contraction)
Myosin motor cycle (5 steps):
Myosin binds actin (tight)
ATP binds myosin → releases from actin
ATP → ADP + Pi → myosin changes conformation (relaxed)
Pi released → myosin re-binds actin (strongly)
ADP released → myosin changes conformation → actin is pulled left (back to step 1)
Cycle repeats with each new ATP during muscle contraction
Each ATP = small movement (few nm) along actin
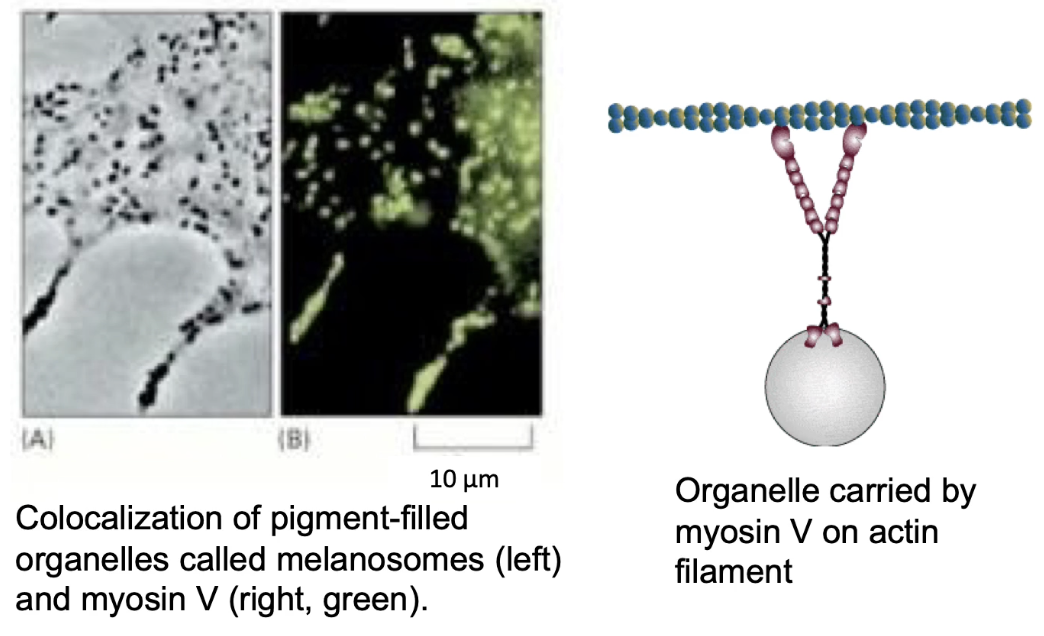
What is the role of Myosin V in cells?
Myosin V powers intracellular cargo transport along actin
Example: moves melanosomes in melanocytes
Melanosomes (membrane-enclosed organelles) = vesicles with melanin pigment
Melanocytes in the skin have dendrites that connect to keratinocytes
Melanin is transferred to keratinocytes → protects DNA from UV (tanning process)
Melanosomes (melanin-filled vesicles) are transported:
By microtubules (long-range)
By Myosin V (final delivery along actin filaments to cell membrane)
Myosin V ensures proper distribution of pigment at the apical surface
Loss of Myosin V function → dilute phenotype in animals
Pigments not properly delivered → lighter/diluted fur color
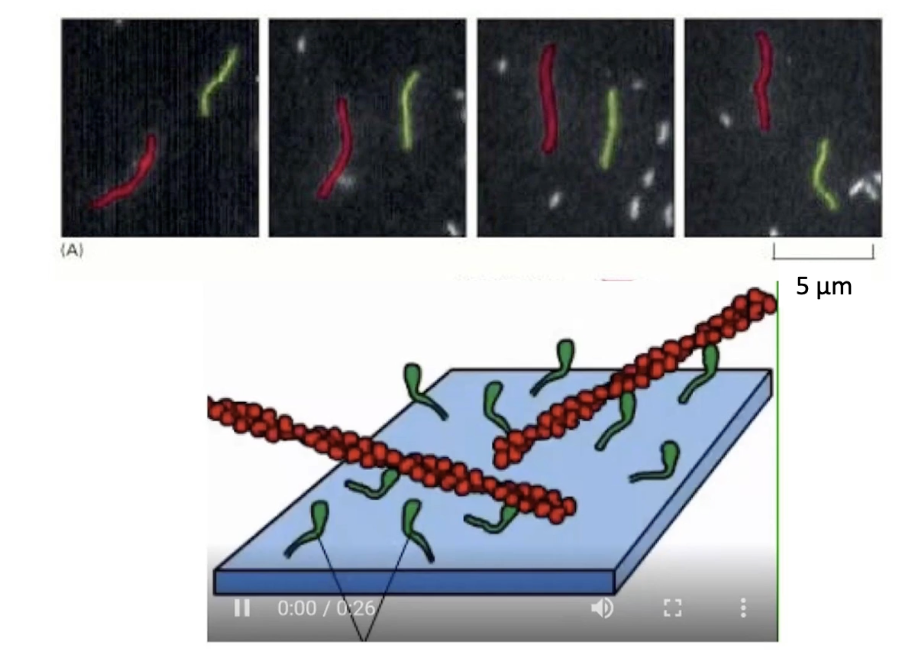
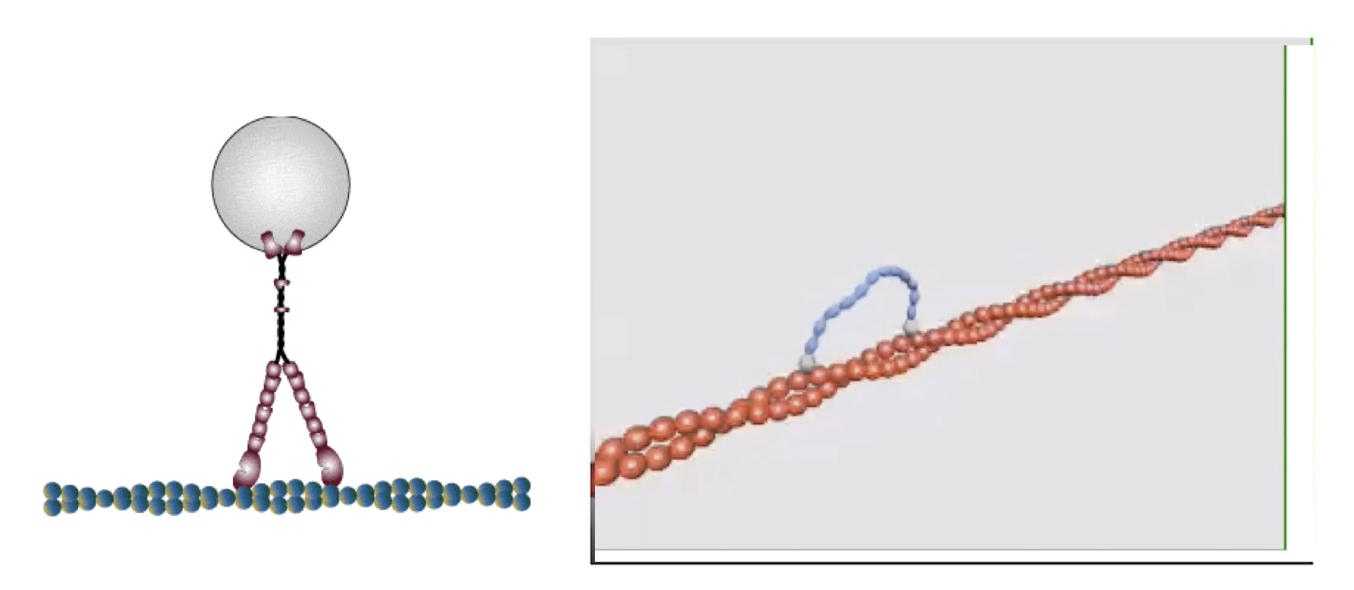
How is myosin movement studied in vitro?
Myosin proteins are fixed (by tails) to microscope slides
Fluorescent actin filaments + ATP are added
Movement of actin = visual indicator of myosin activity
ATP cycle (binding → hydrolysis → Pi/ADP release) powers motion
Movement is caused by myosin motor heads cycling through conformations
Seen under a microscope as fluorescent actin motion
What affects the speed of different myosin proteins?
Myosin speed = 0.2 to 60 µm/sec, varies by type
Influenced by:
ATPase rate (how fast ATP is hydrolyzed in myosin head)
Actin binding time (how long myosin stays bound due to affinity)
Myosin II:
Binds actin only ~5% of the cycle → faster movement
Myosin V:
Binds actin ~90% of the cycle → slower but stable movement
What determines the step size of myosin, and how does myosin V move along actin filaments?
Step size of myosin depends on lever arm length
Longer lever → bigger step during power stroke
Myosin V lever is 3× longer than myosin II
Step size:
Myosin II: ~7 nm
Myosin V: ~36 nm
Myosin V movement:
Moves in hand-over-hand fashion
Trailing head detaches → swings forward → becomes leading head
Moves toward the barbed/plus end of actin filament
Efficient for cargo transport due to large steps and strong actin binding
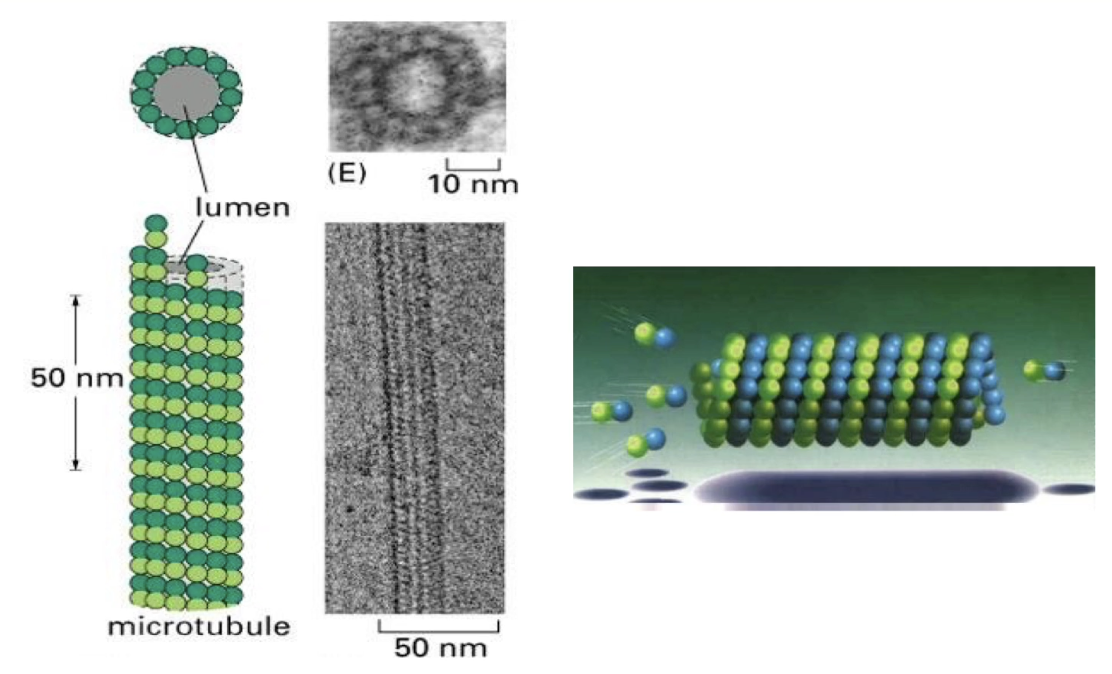
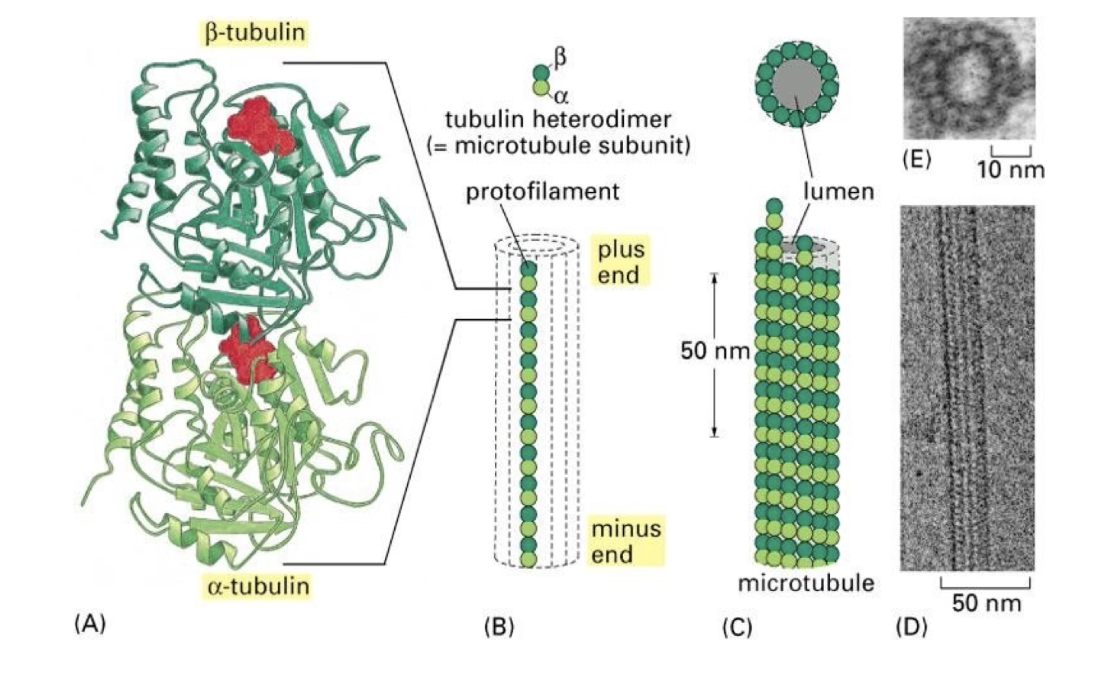
What is the structural organization of microtubules?
Microtubules = hollow tubes made of 13 protofilaments
Protofilaments arranged in a circular pattern → strong tube wall
Basic unit = α/β-tubulin dimers
Protofilaments are staggered → dimers appear to spiral like a spring
Structure visible via electron microscopy
How do α and β tubulin subunits interact with GTP?
Both α & β tubulin bind GTP
α-tubulin: GTP is tightly bound, never hydrolyzed or exchanged
β-tubulin: GTP is hydrolyzed to GDP, then exchanged back to GTP
Tubulin dimers are added/removed as α-β pairs
α-β-GTP dimers: high affinity for microtubules
α-β-GDP dimers: low affinity → more likely to dissociate
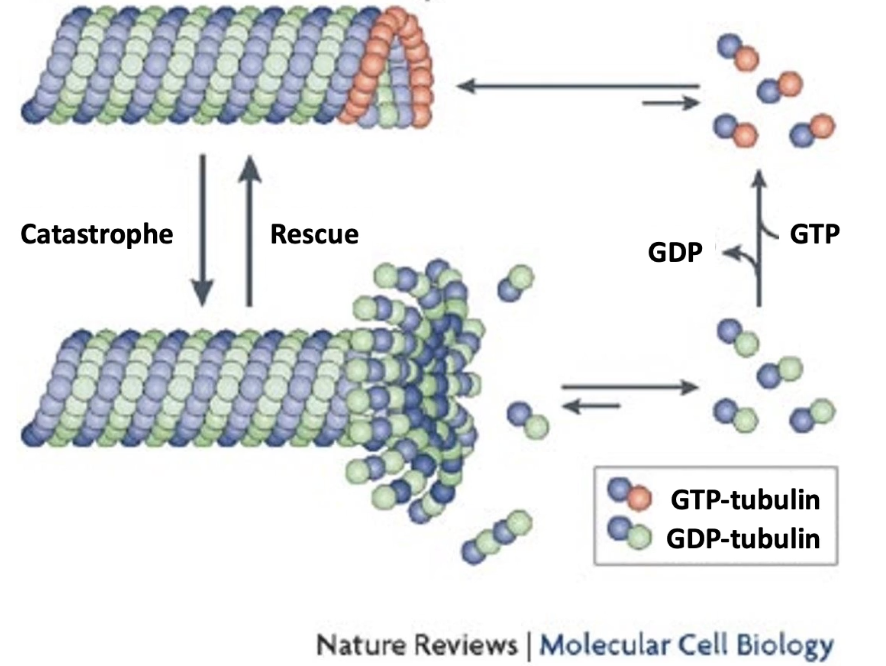
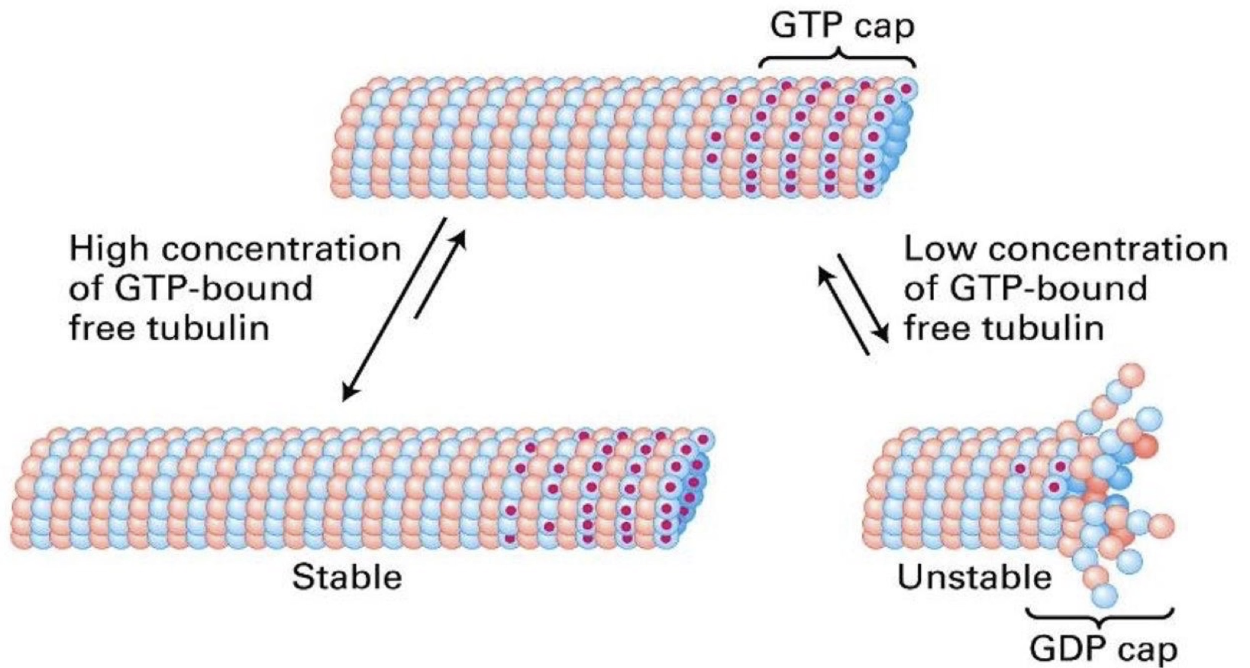
How are microtubules polarized and what drives their growth/shrinkage?
Microtubules are polar:
Plus-end: fast-growing
Minus-end: slow-growing
Dimer orientation:
β-subunit toward plus-end
α-subunit toward minus-end
Growth ("rescue"): α-β-GTP dimers added to plus-end
Shrinkage ("catastrophe"): α-β-GDP dimers released
GTP hydrolysis happens after polymerization → most of microtubule = α-β-GDP
GTP cap (or α-β-GTP) on plus-end promotes growth; loss of cap → shrinkage
4× slower dissociation rate than α-β-GDP
Due to higher affinity between α-β-GTP dimers and their neighbors
What is EB1 and how does it affect microtubule growth?
EB1-GFP is a plus-end binding protein
Prevents premature catastrophes
Acts as a positive regulator of microtubule growth
Visualized in live cells using RFP-tubulin (red) and EB1-GFP (green)
Localizes specifically to growing plus-ends of microtubules
What is dynamic instability in microtubules?
Plus-end undergoes oscillation between growth and shrinkage
Growth = polymerization = rescue
Shrinkage = depolymerization = catastrophe
Maintained by concentration of free α-β-GTP dimers that allows polymerization
Ensures constant remodeling of microtubule structure
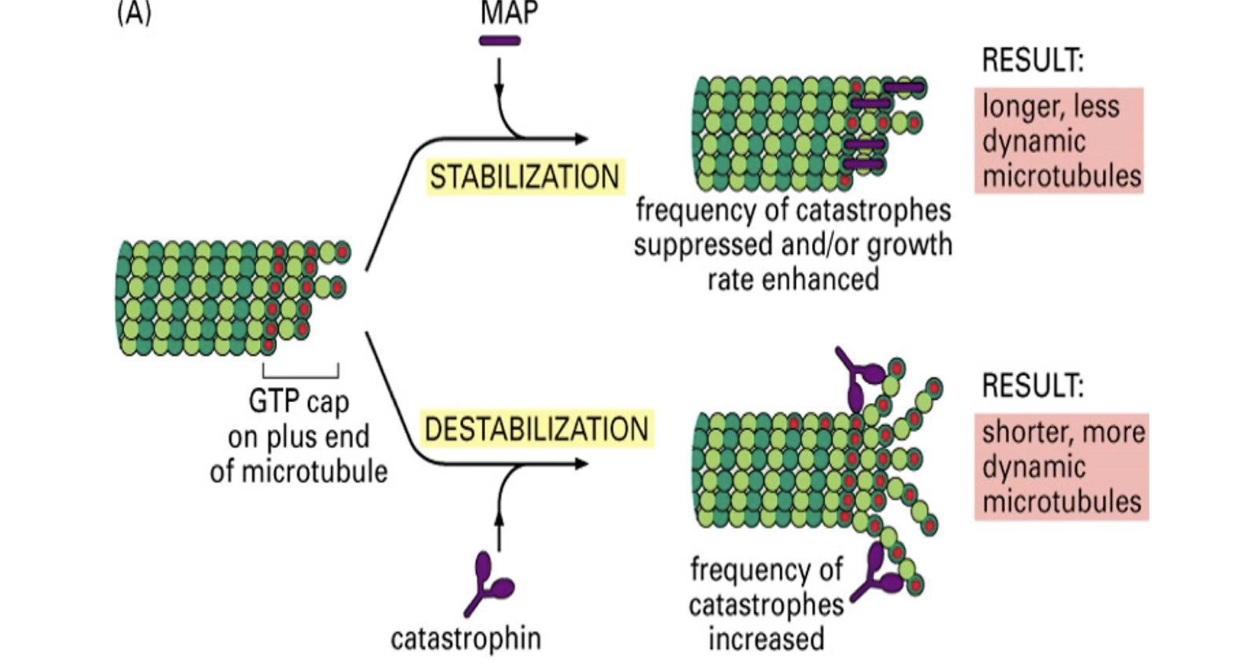
What are MAPs and what roles do they play in microtubule function?
MAPs = proteins that regulate microtubule assembly/disassembly
Functions:
Bundle formation (cross-linking)
Stability and rigidity control
Regulation of assembly rate
Two functional groups:
Stabilizers: Tau, EB1
Destabilizers: Catastrophin
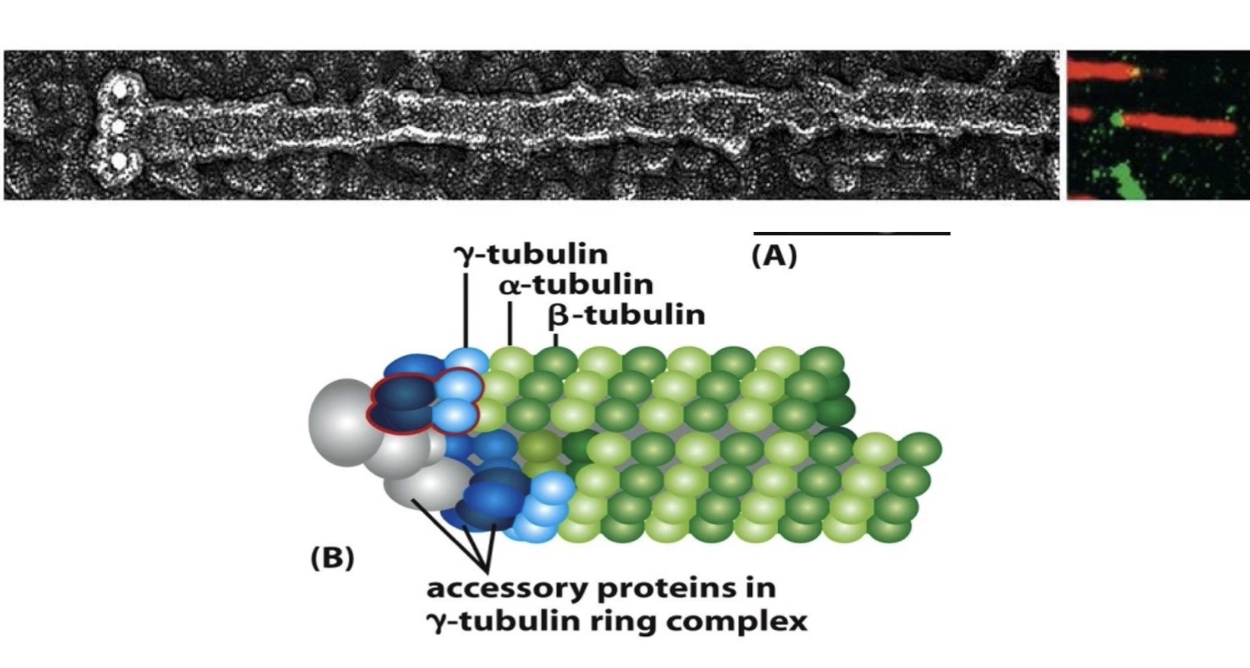
What role does γ-tubulin play in microtubule formation?
γ-tubulin = nucleation factor (not part of microtubule body)
Present in small amounts
Forms γ-TuRC (γ-tubulin ring complex) with other proteins
Nucleates minus-end of microtubule
Acts as a template for plus-end growth
Caps the minus-end → growth occurs only at the plus-end
What is the MTOC and how does it organize microtubule growth?
MTOC = Microtubule Organizing Center
In animal cells → called centrosome, located near the nucleus
Contains:
Two centrioles
Pericentriolar material (PCM) with γ-TuRC complexes
γ-TuRC nucleates minus-ends
Plus-ends grow outward toward cell periphery
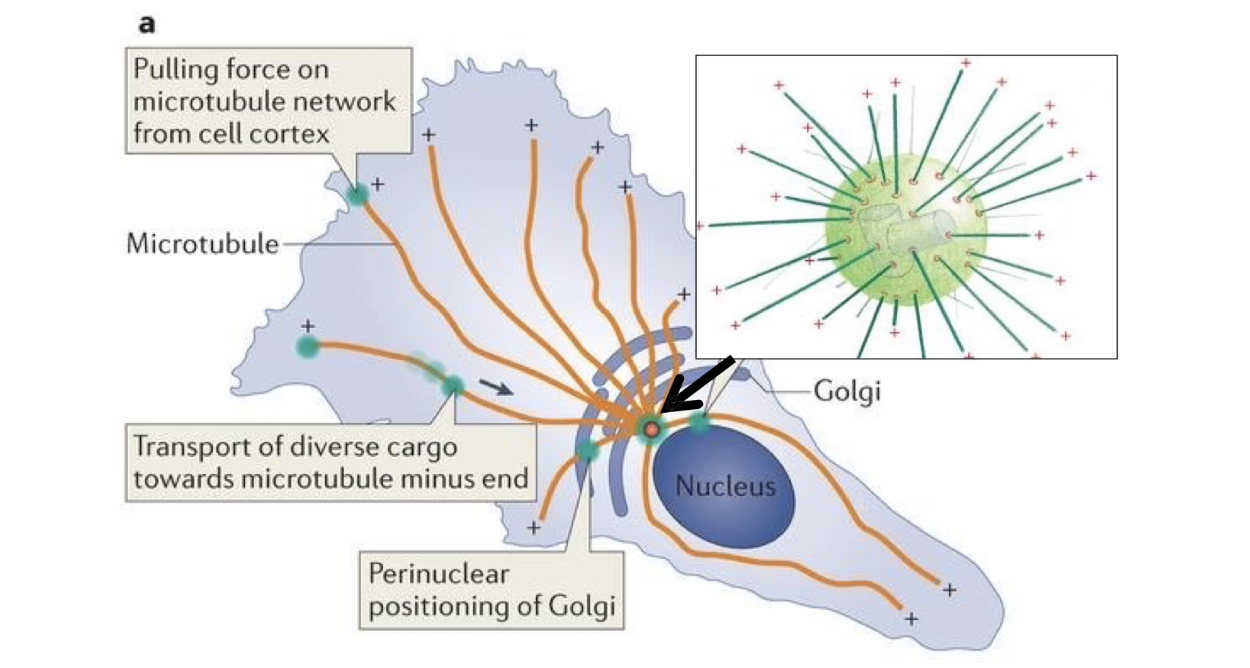
What is the role of the MTOC during mitosis?
MTOC = Microtubule Organizing Center, duplicates in mitosis
Forms the mitotic spindle
Microtubules nucleated from γ-TuRC at each MTOC
Plus-ends grow outward
Some anchor the spindle to cell cortex
Others extend inward to attach to chromosomes
Microtubules guide sister chromatid separation
Spindle is dynamic – built and disassembled based on microtubule instability
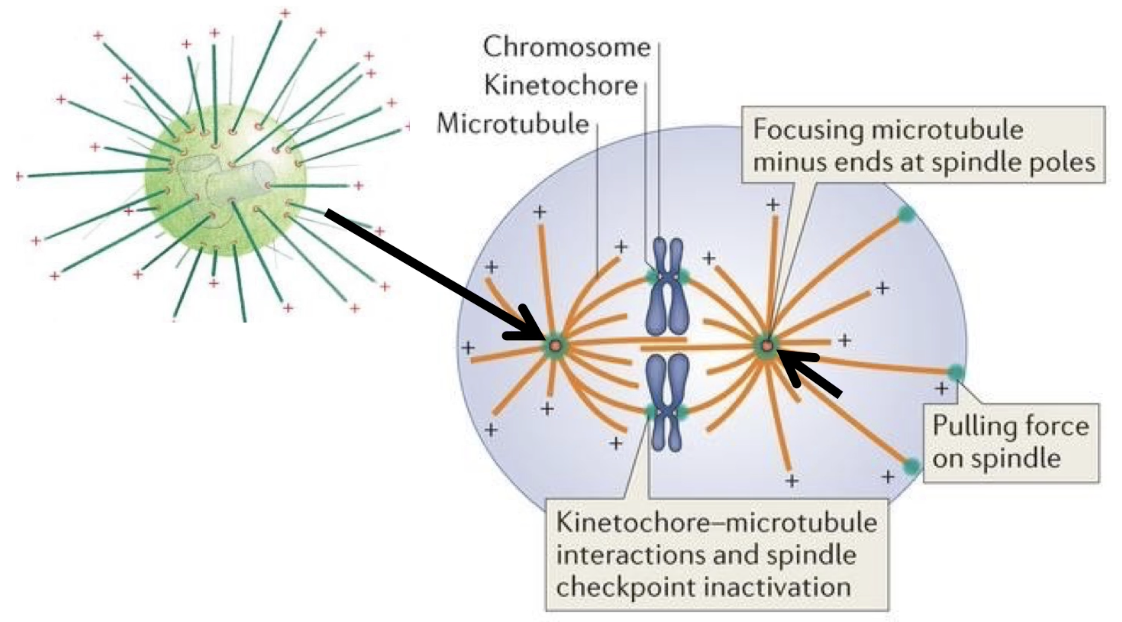
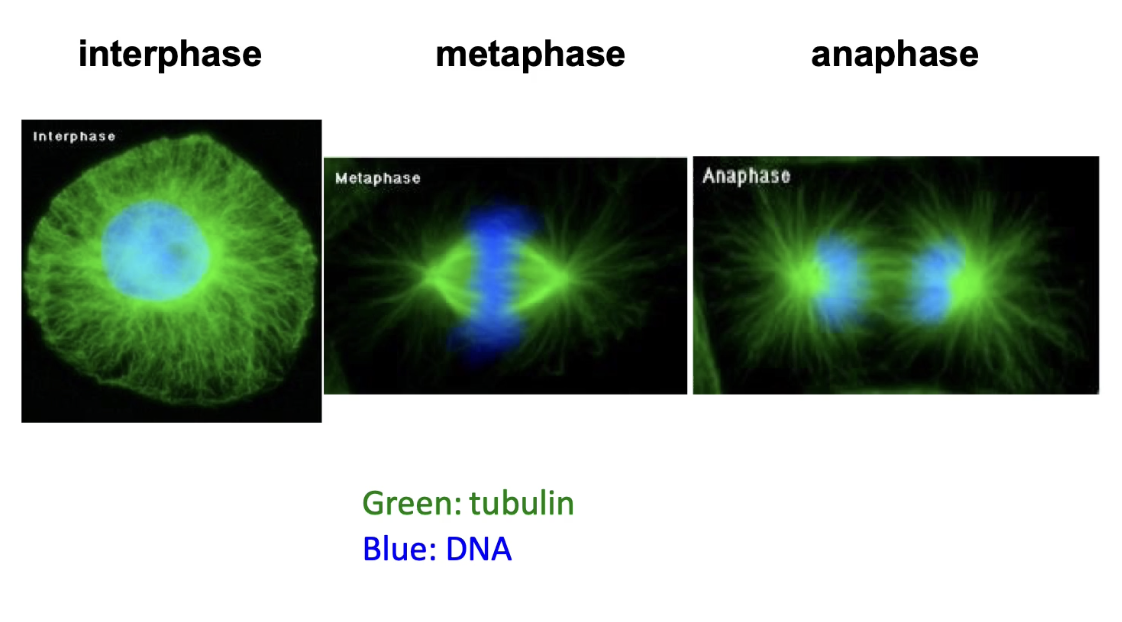
How do microtubules assist in chromosome movement during mitosis?
Tubulin visualized via antibodies or GFP constructs
DAPI stains DNA (nucleus = blue)
In interphase: microtubules fill the cytoplasm
In metaphase:
Replicated chromosomes align at the spindle equator
Spindle microtubules attach to centromeres
In anaphase:
Spindle poles move apart
Microtubules shorten → chromatids pulled to poles
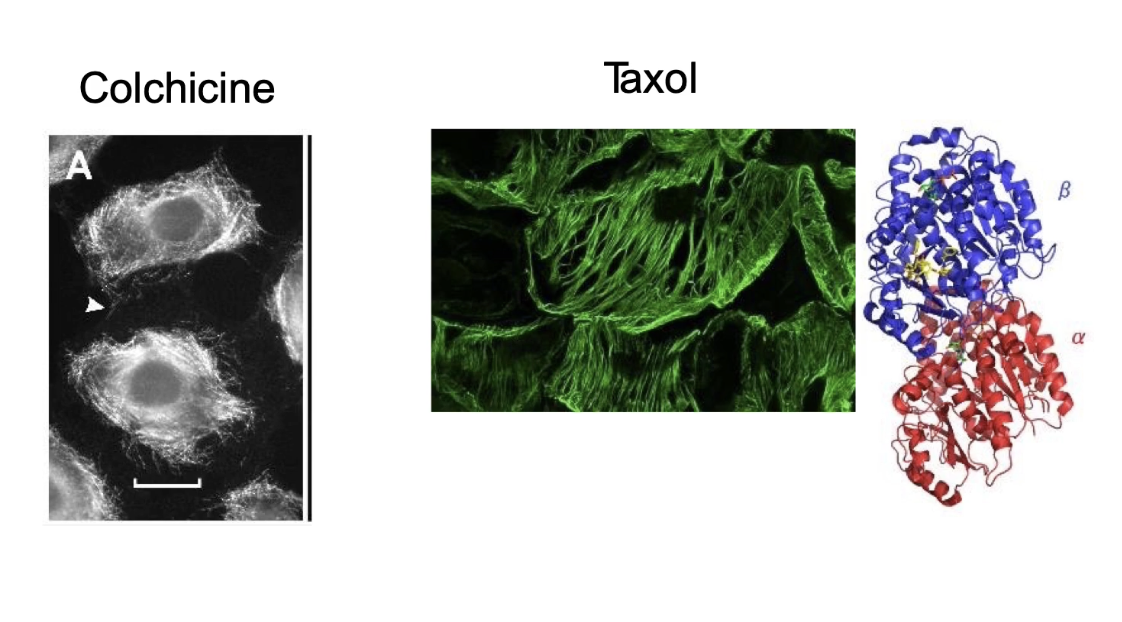
What drugs affect microtubule dynamics and how do they work?
Colchicine (meadow saffron or autumn crocus):
Inhibit polymerization.
Binds to free αβ-tubulin dimers.
Bound dimers can still join a growing microtubule.
BUT they block further addition or removal of tubulin → microtubules can't grow or shrink.
Effect on mitosis: Cells arrest in metaphase (chromatids can't separate).
Taxol (Paclitaxel):
Binds to β-tubulin in the microtubule.
Increases stability by preventing depolymerization (shrinkage).
Effect on mitosis: Microtubules are too stable → spindle can't form properly, mitosis is inhibited.
Used as a cancer treatment (from Pacific yew tree).
Vinblastine & Nocodazole:
Cause rapid depolymerization of microtubules.
Disrupt spindle formation → inhibit mitosis.
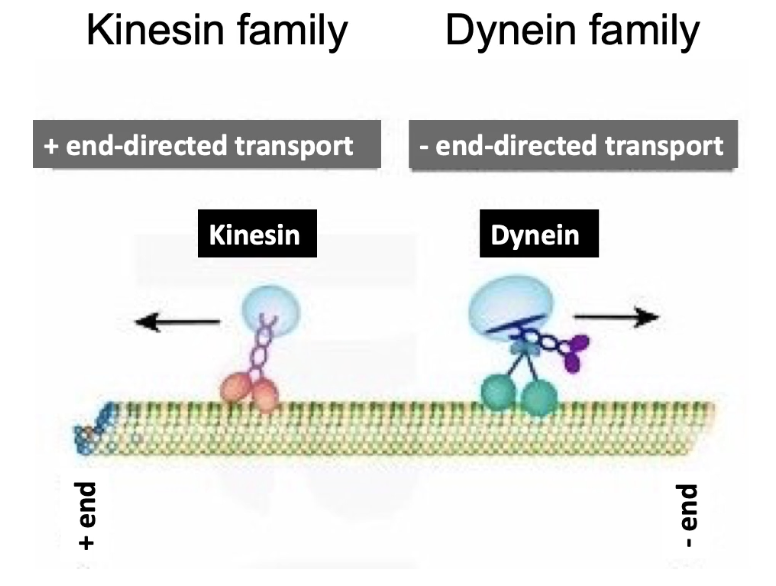
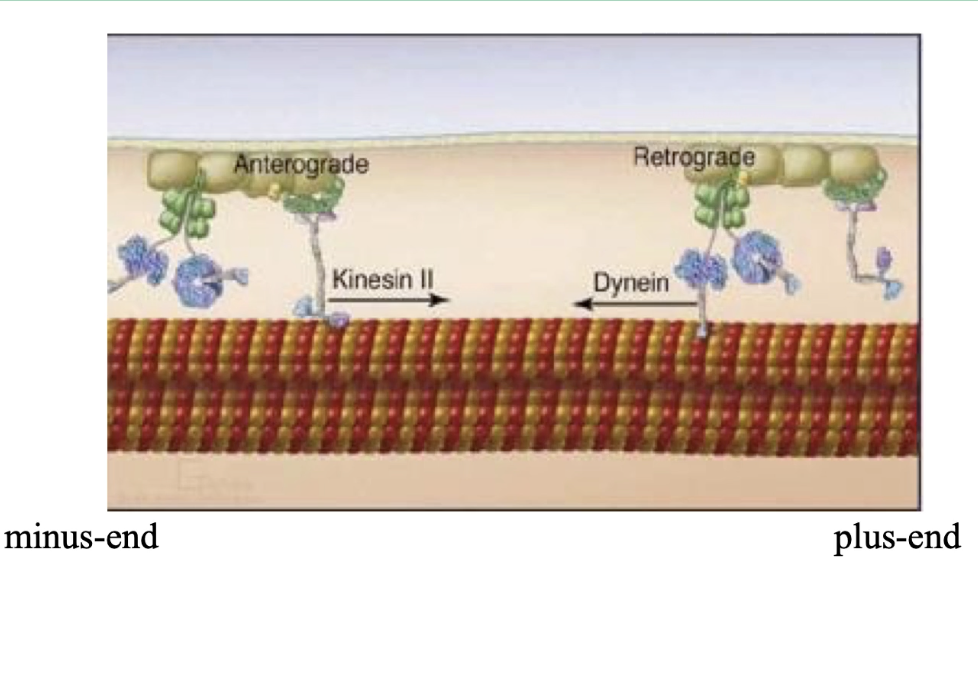
What are the two main types of motor proteins that move along microtubules, and in which directions do they move?
Kinesin:
Moves toward the plus end of microtubules
Transports cargo toward the cell periphery
Dynein:
Moves toward the minus end of microtubules
Transports cargo toward the cell center/MTOC
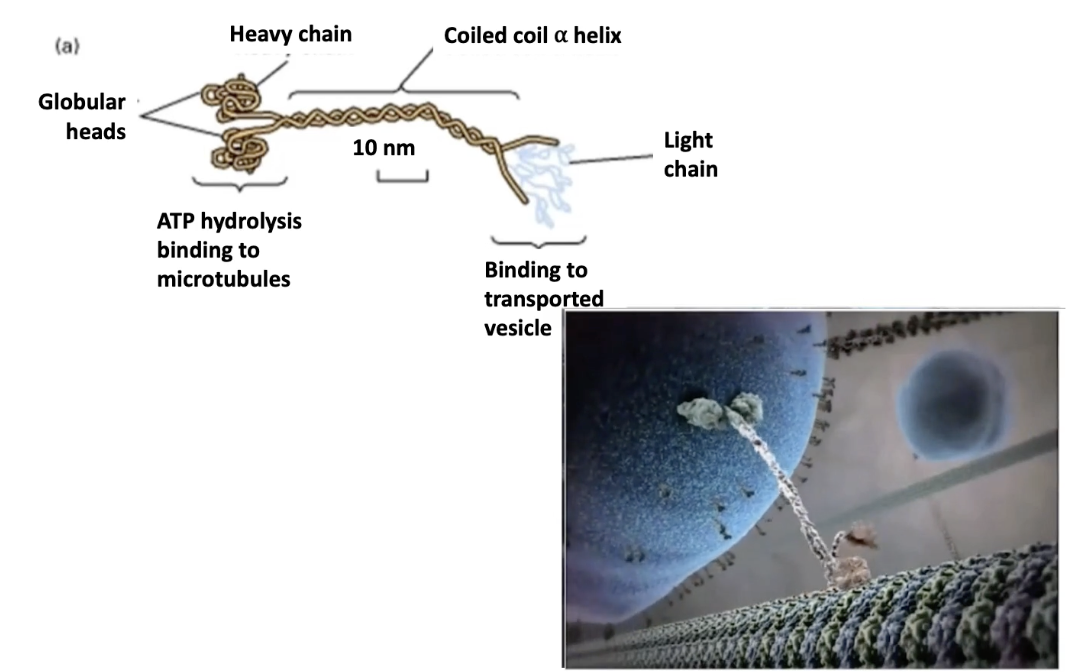
What is the structure of kinesin, and how does it function?
Structure:
Tetramer = 2 heavy chains + 2 light chains
N-terminal globular motor domains bind microtubules
Heavy chains = motor activity
Light chains = cargo binding (via variable tails)
Function:
Motor domains use ATP hydrolysis for movement
Transports vesicles and organelles to plus ends of microtubules
Moves cargo away from MTOC → toward cell edge
Tail regions determine cargo specificity
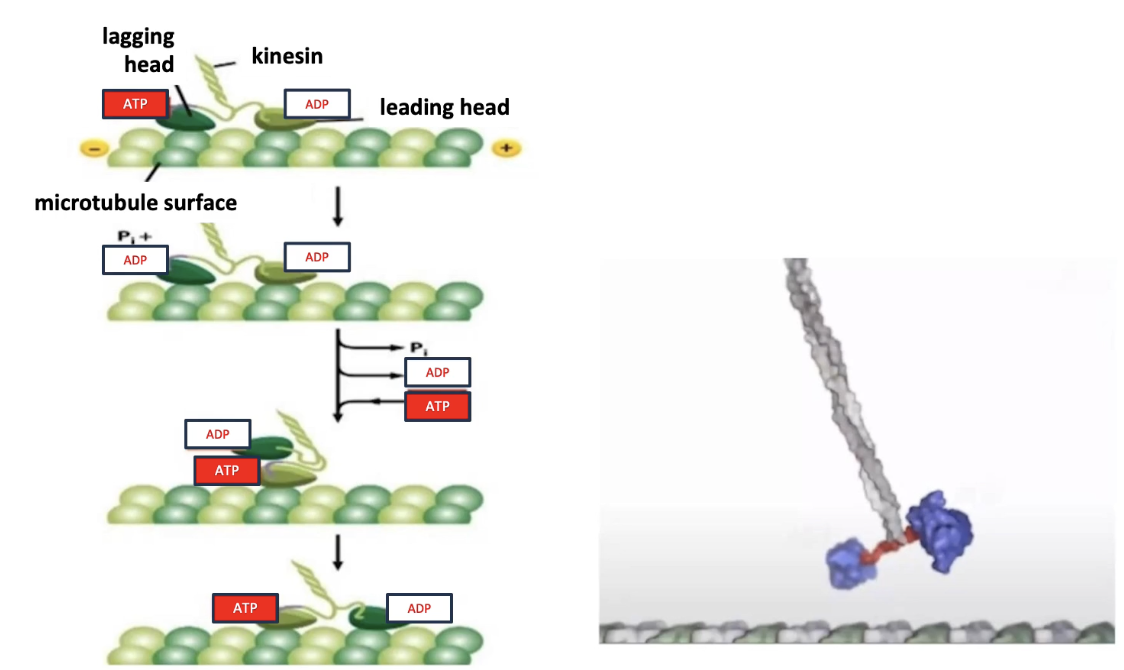
How does kinesin move along microtubules in a “hand-over-hand” fashion?
Mechanism:
Two motor heads (dimers) take turns moving
At least one head always remains attached to the microtubule
Cycle stages:
Lagging head: bound to ATP → hydrolyzes ATP → ADP + Pi → releases microtubule
Leading head: bound to ADP → exchanges for ATP → tightens microtubule binding
Conformational change in neck swings lagging head forward
This resets the cycle for continuous forward movement
Key concepts:
Movement is ATP-driven
Coordination between heads ensures continuous stepping
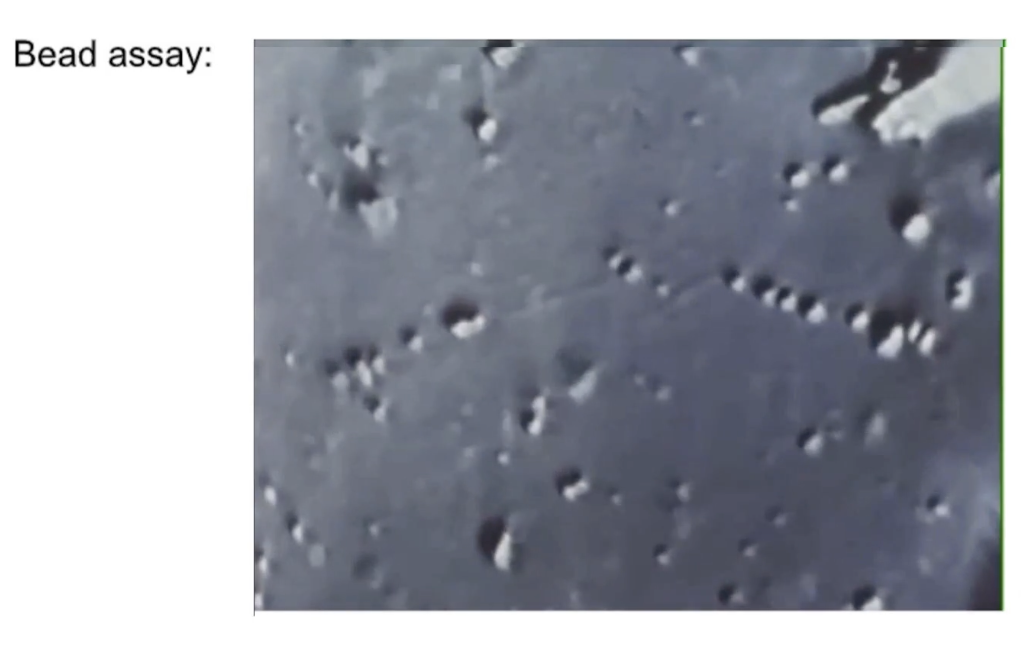
How can kinesin movement be studied using plastic beads in vitro?
Nomarski microscope assay:
Plastic beads (1 μm) tethered to kinesin
Kinesin walks along a microtubule fixed to a dish
Microtubules made of purified tubulin
Bead motion visualized in real time
Movement rate: 0.5 μm/second
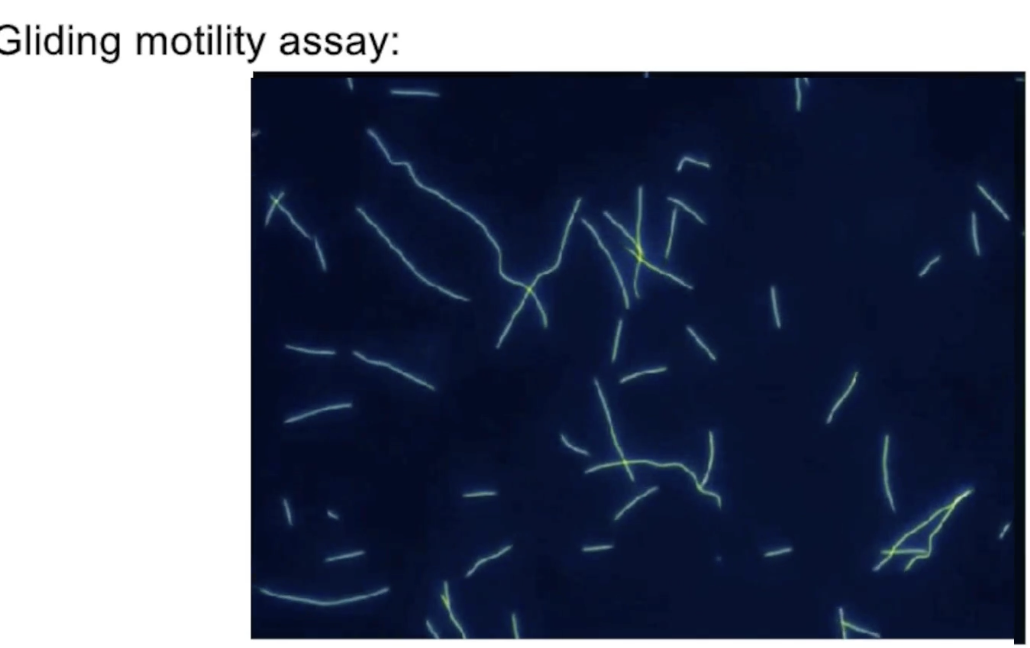
What is a gliding mobility assay and how does it measure kinesin movement?
Setup:
Kinesin proteins are anchored to a glass slide by their tails
Fluorescent microtubules are added to the solution
Action:
Kinesins move the microtubules across the slide
Microtubule movement is visualized with a fluorescence microscope
What is dynein and how does it function in cells?
Dynein = minus-end directed motor
Moves toward MTOC, away from cell edge
Two types:
Cytoplasmic dynein: moves organelles and vesicles
Axonemal dynein: powers cilia and flagella movement (e.g., sperm cells)
Structure:
2 identical heavy chains
Multiple intermediate + light chains
Movement:
ATP hydrolysis enables movement (described as a “strange walk”)

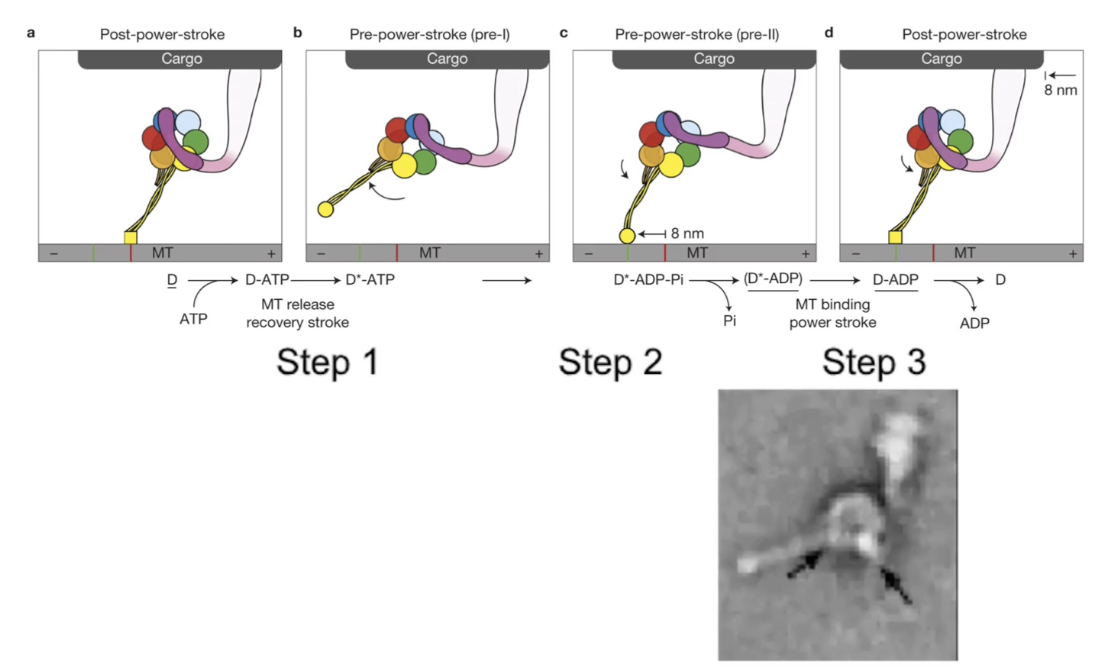
How does dynein move cargo using its power-stroke mechanism?
Steps of dynein power-stroke:
ATP binds → motor head releases microtubule
ATP hydrolysis → dynein-ADP+Pi reattaches to microtubule
Pi release → triggers power-stroke via linker arm (moves cargo)
Within a dimer:
Dyneins alternate power-strokes for continuous movement
Each stroke moves cargo ~8 nm toward minus end
Electron microscopy:
Confirms visual progression of power-stroke steps
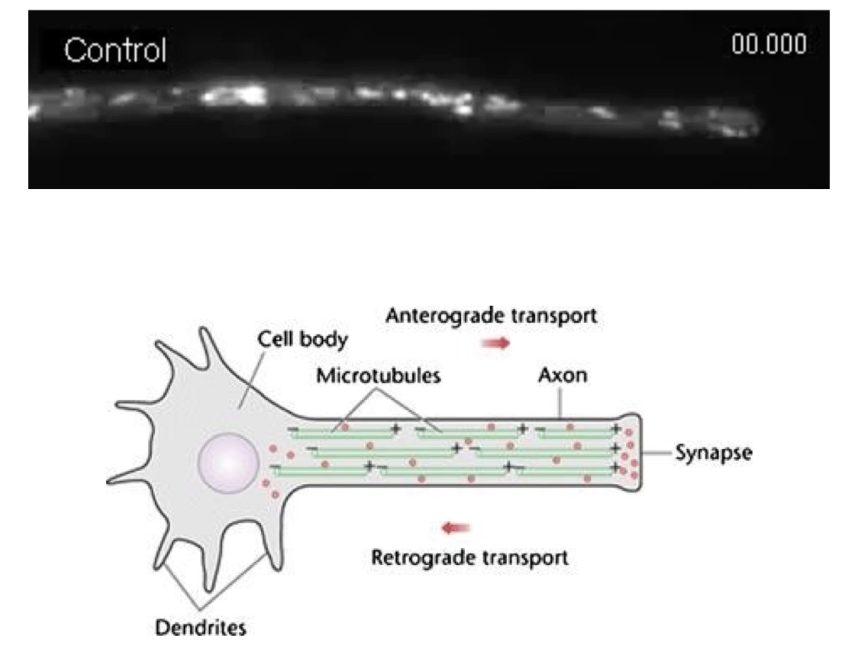
How do motor proteins transport cargo along microtubules in cells like neurons?
Cargo uses both kinesin and dynein motor proteins
Movement direction depends on which motor is active
Minus-end anchored at MTOC near the nucleus
Plus-end extends toward cell membrane/synapse
Example: Neurotransmitter vesicles move along axons
From cell body ➝ synapse
Can move in both directions on microtubule “highways”
How is cargo direction decided when two motor proteins pull in opposite directions?
Dynein pulls to minus-end, kinesin to plus-end
"Tug-of-war" model:
Motors compete to pull cargo
Final direction = winner of the battle
Regulatory proteins control which motor is active
Respond to internal cellular signals
What is an example of a single vesicle reversing direction on a microtubule?
Fluorescently-labeled vesicle tracked in cell
Initially moves toward minus-end
In 9th frame, reverses direction to move toward plus-end
Shows dynamic, bidirectional transport on microtubules
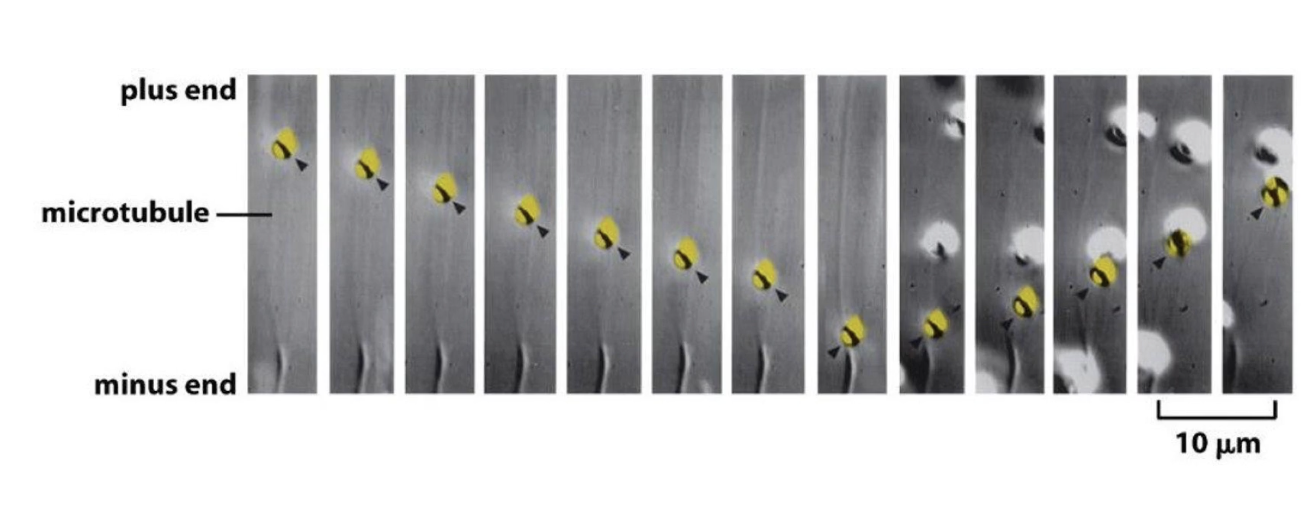
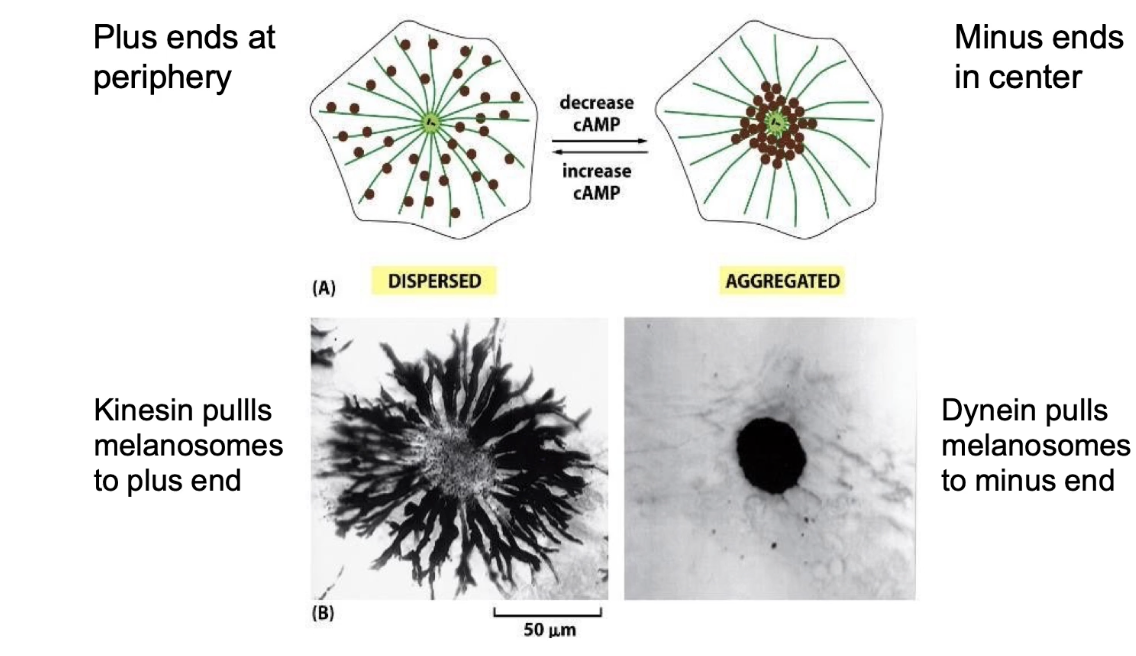
How do molecular motors regulate melanosome movement and skin color in fish?
Melanosomes = pigment-filled organelles
Transport affects skin color change in response to signals
Dynein:
Moves melanosomes to center (minus-end)
Cell appears lighter
Kinesin:
Moves melanosomes to periphery (plus-end)
Cell appears darker
Switch between directions controlled by:
cAMP as a secondary messenger in signal transduction pathways
Applied Lecture
Cytoplasmic streaming in plants
What is cytoplasmic streaming?
Directed flow of cytosol and organelles around cells, especially prevalent in plant cells (can be seen in fruit fly and c.elegan embryos), aiding nutrient and metabolite delivery.
Example: Chloroplast movement in Elodea.
Who first reported cytoplasmic streaming and when?
Bonaventura Corti in 1774, studying Nitella and Chara green algae
Occurs from algae to angiosperm (higher order) flowering plants
Seen more in aquatic plants > land plants
Primitive + essential
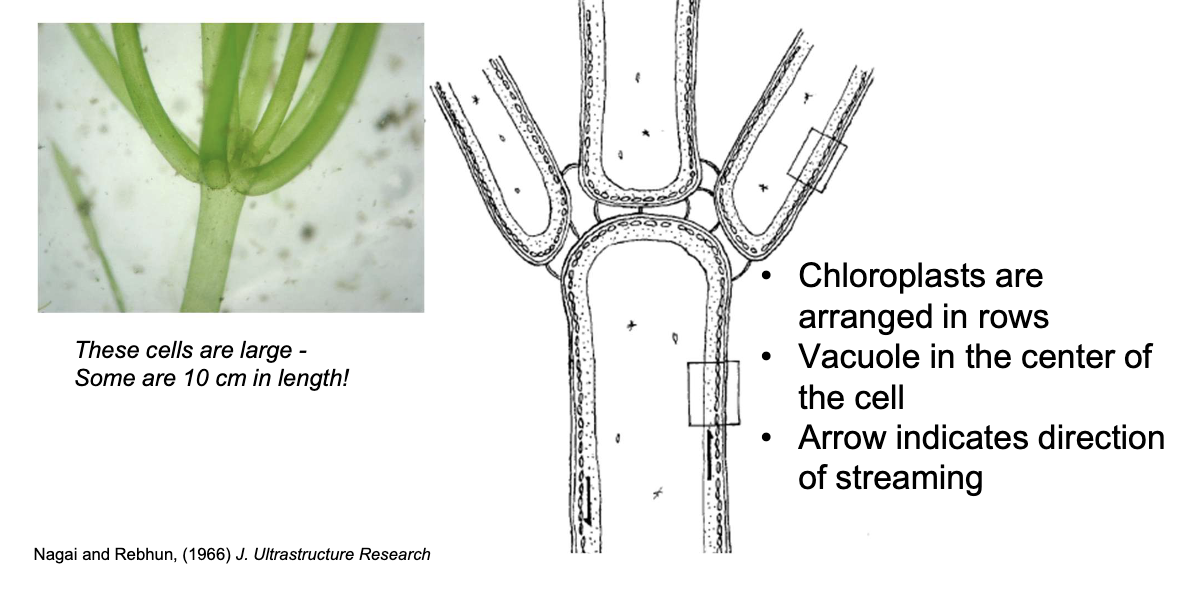
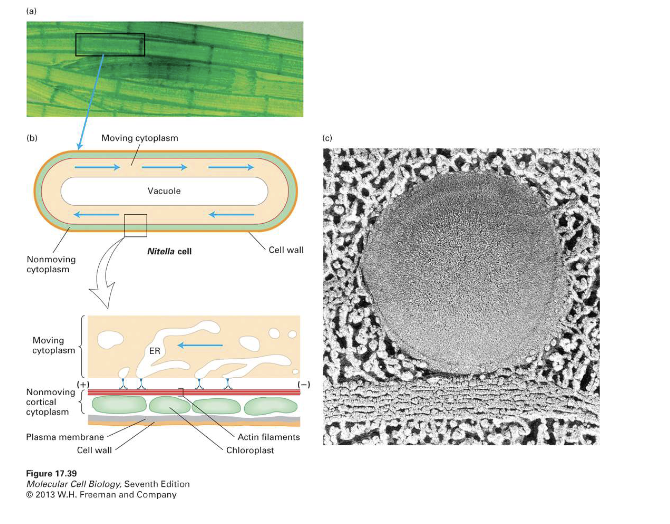
What cell structures are involved in rotational streaming in Nitella?
Internodal and leaf cells
Actin filaments near the cell membrane at these regions, chloroplasts arranged in rows, and a central vacuole
Function: Circling chloroplasts help make energy for aquatic plants, where light sources are not as available.
What composes the microfilament bundles in internodal cells and their suggested identity?
Each microfilament is composed of twisted subunits, suggested to be actin filaments.

How does cytochalasin affect cytoplasmic streaming?
A fungal metabolite that binds actin filaments and blocks polymerization, showing streaming velocity depends on actin microfilaments.
Control: DMSO since cytochalasin cannot be injected directly
Experimental Condition: DMSO + cytochalasin
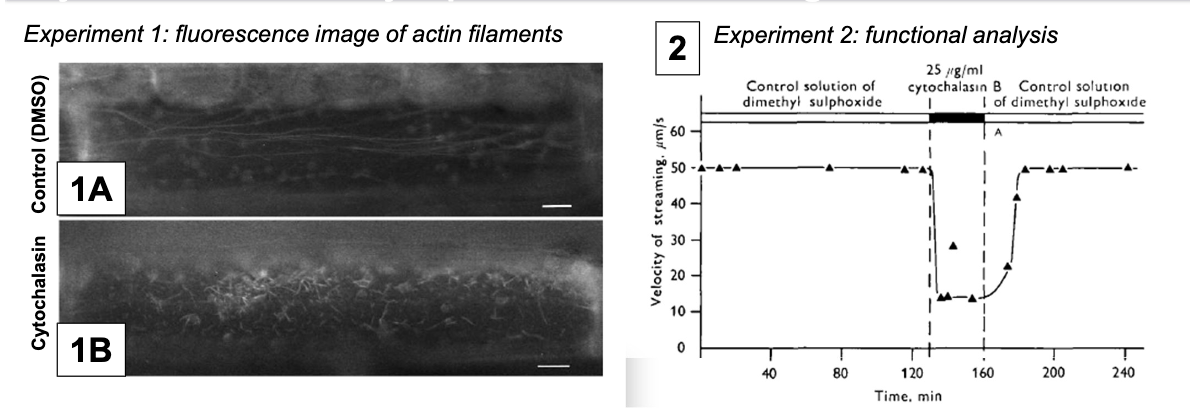
How is cytoplasmic streaming powered?
Myosin motor proteins move along actin filaments using ATP hydrolysis, enabling high velocities (up to 70 μm/s in Chara algae)
Good for large cells diffusion is slow
High velocity due to high ATPase activity and fast ADP dissociation
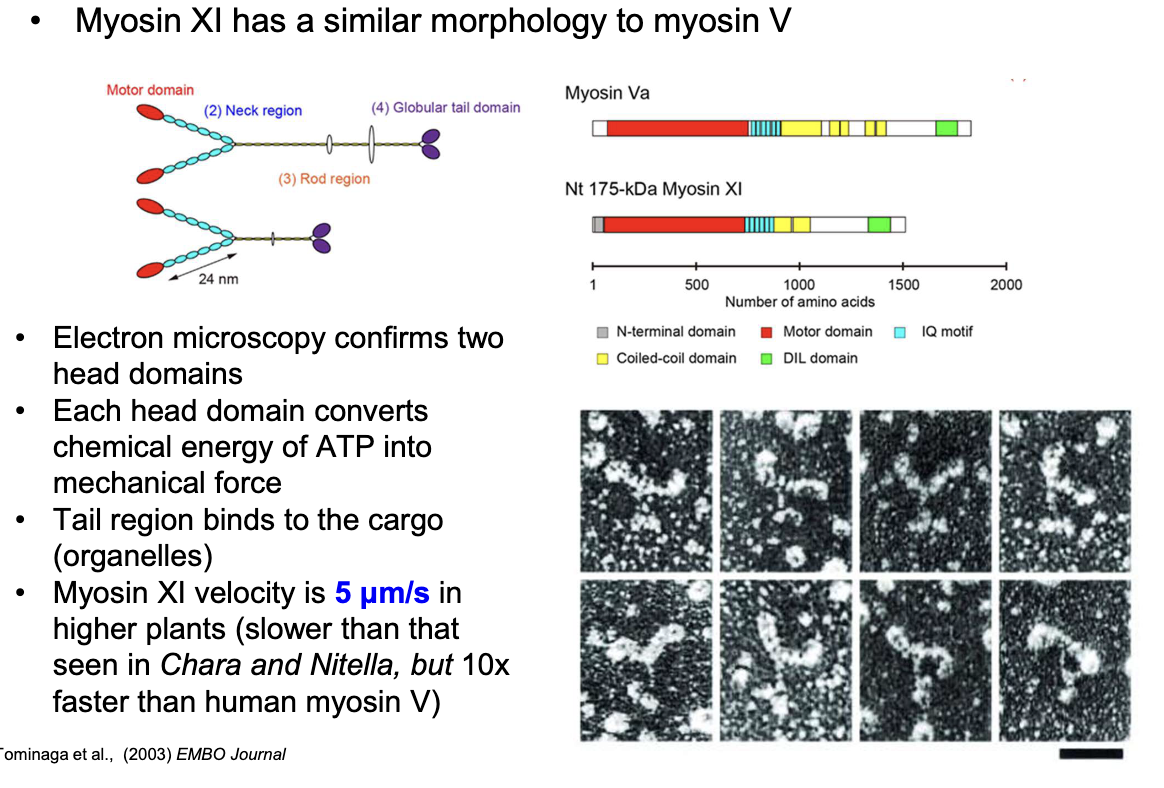
What is special about myosin XI (higher order plants)?
Similar structure to Myosin V (humans)
It has two ATP-converting head domains and a tail binding cargo
Moves at 5 μm/s (10x faster than human myosin V, but slower than myosin in Chara and Nitella plants)
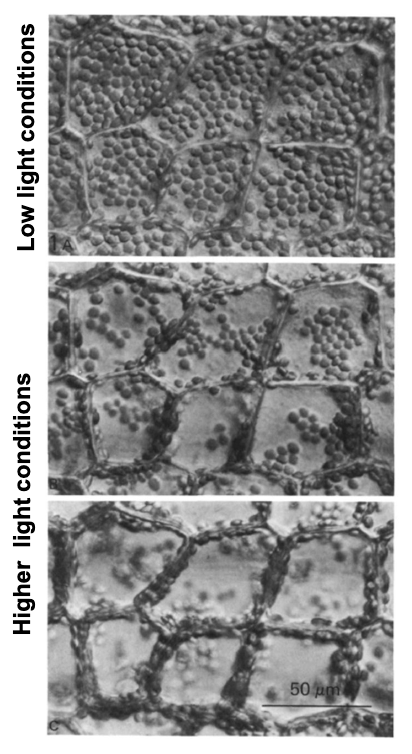
What influences cytoplasmic streaming in plants and algae?
Streaming is usually constant (primary streaming) but can be induced in higher aquatic plants by light or chemicals that change ATP availability, affecting chloroplast position.
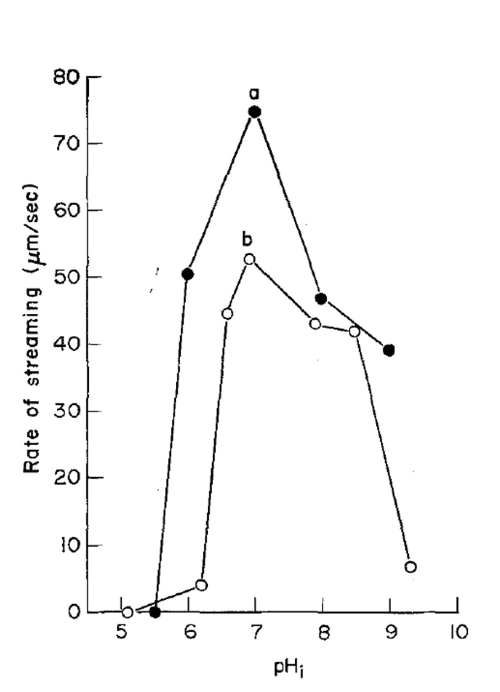
How does pH affect cytoplasmic streaming rate?
Maximal streaming occurs at pH ~7, with decreased rates in acidic or alkaline conditions.
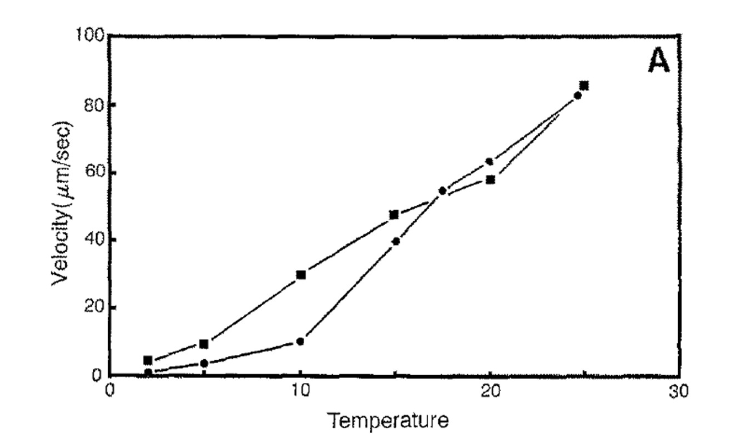
How does temperature affect cytoplasmic streaming velocity?
Velocity increases linearly with temperature.
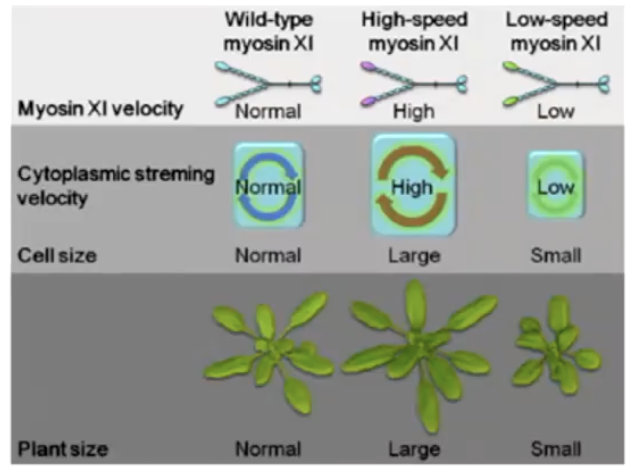
What happens when myosin XI is knocked out in plants?
Growth defects, reduced cell size, and delayed flowering, suggesting cytoplasmic streaming is important for development.
What was done to study cytoplasmic streaming effects on plant development?
Motor domains of Arabidopsis myosin XI were replaced with fast (Chara) or slow (human myosin Vb) myosins.
*Myosin XI is same in algae and higher order plants, but velocity differs.
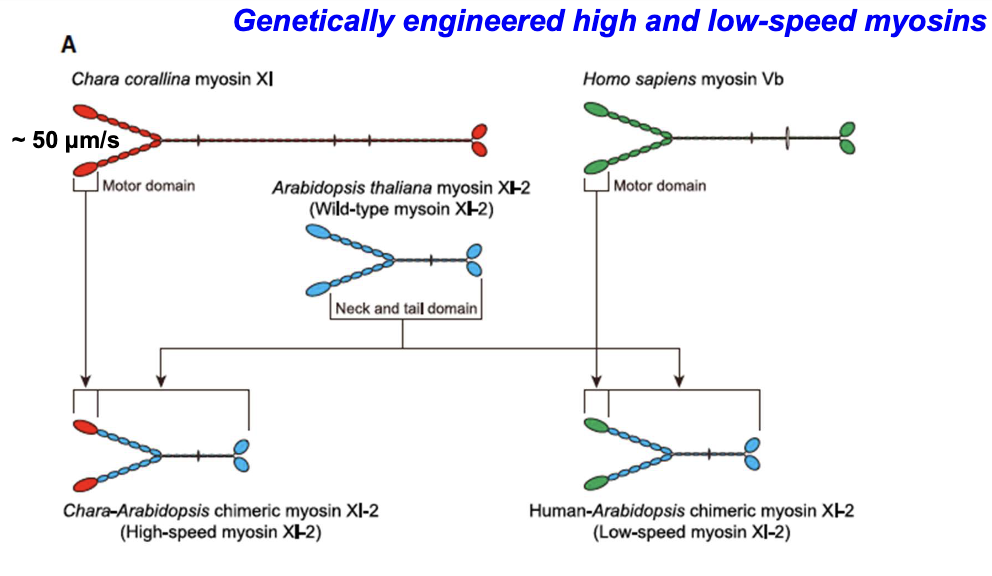
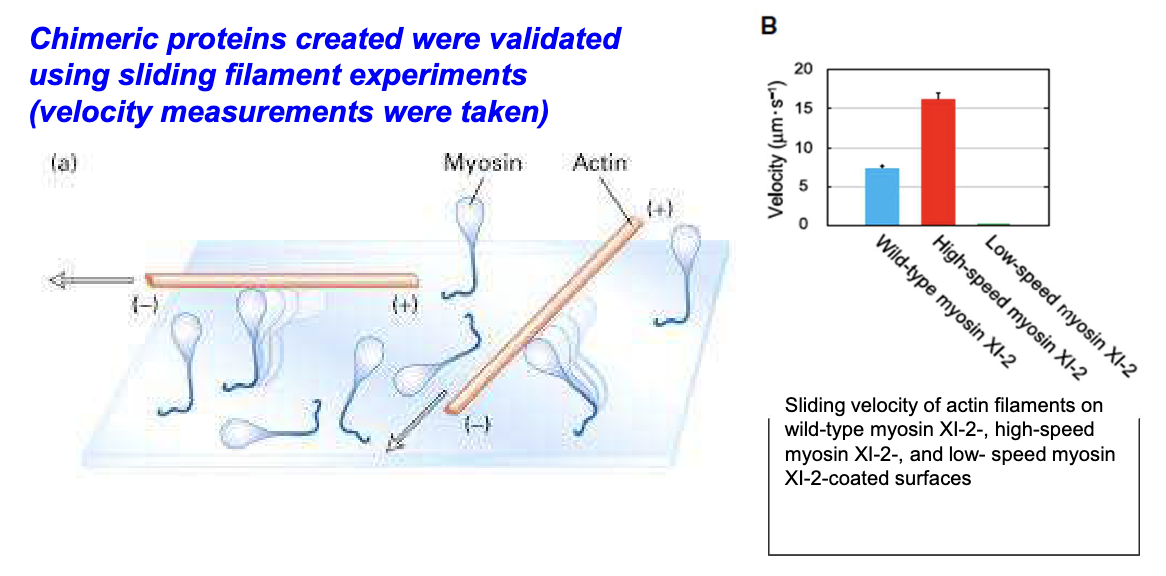
How were chimeric myosins validated?
Using in vitro sliding filament assays to measure velocity; transformed plants carried organelles normally but at altered speeds.
How did high-speed myosin XI affect Arabidopsis growth?
Plants grew taller with larger leaves compared to wild-type and low-speed myosin plants
Shoot height + leaf size + dry weight of modified plants differed from wild type 35 days after planting
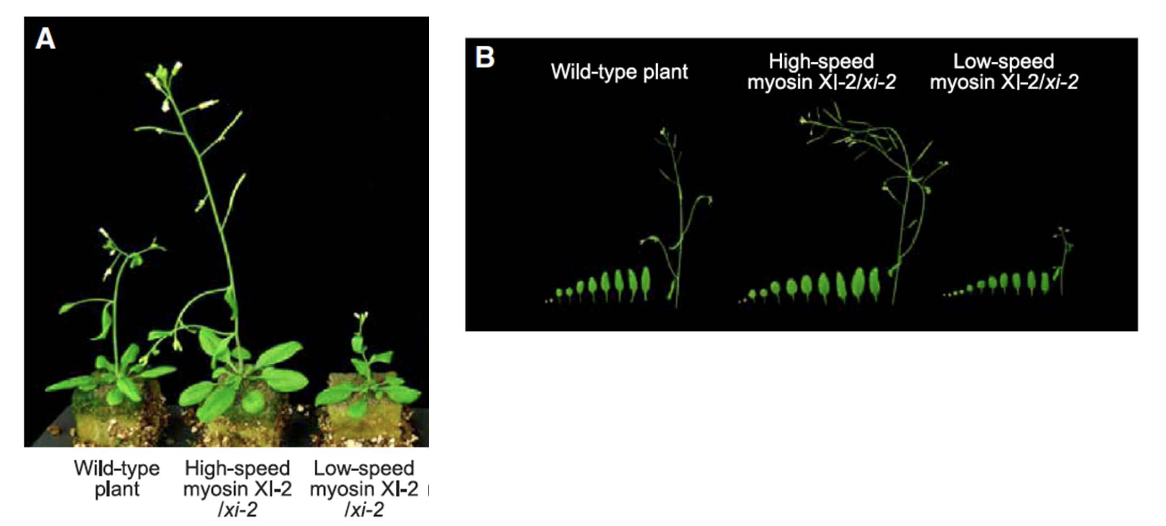
Did chimeric myosins affect leaf number?
No change in leaf number, but high-speed myosin plants had greater shoot height and dry weight.
*Significant stars are comparing modified to wild type
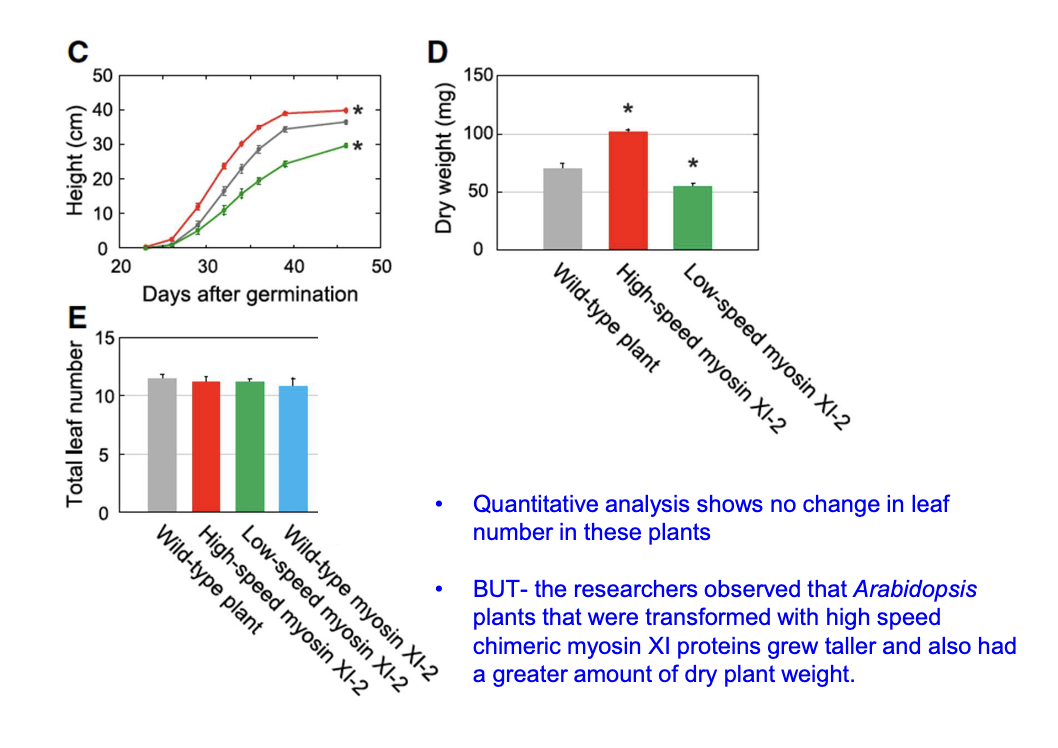
How did cell size change with high-speed and low-speed myosin XI?
High-speed myosin XI cells were 47% larger; low-speed myosin XI cells were 20% smaller than wild-type
No significant difference in number of cells
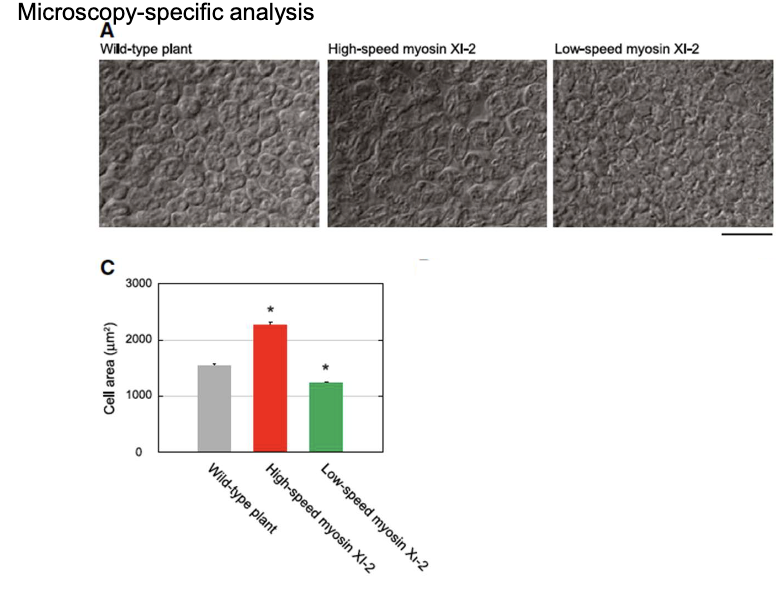
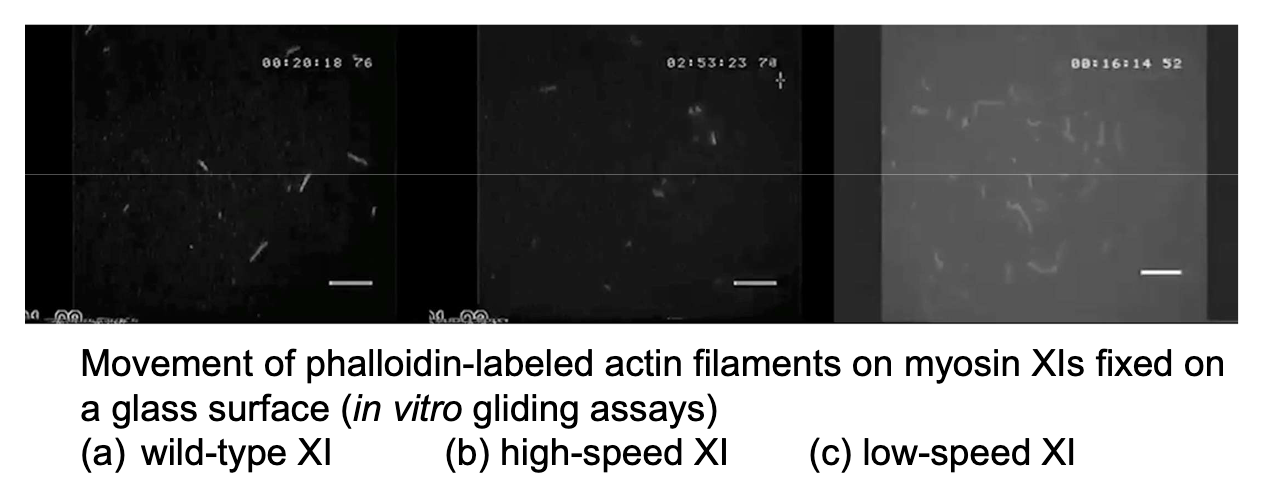
What did in vitro gliding assays reveal about myosin XI variants?
Wild-type, high-speed, and low-speed myosins have distinct velocities affecting filament movement.
How does myosin XI speed affect cytoplasmic streaming in plants?
High-speed myosin XI plants show the fastest cytoplasmic streaming due to faster myosin motor domain “walking” on actin.
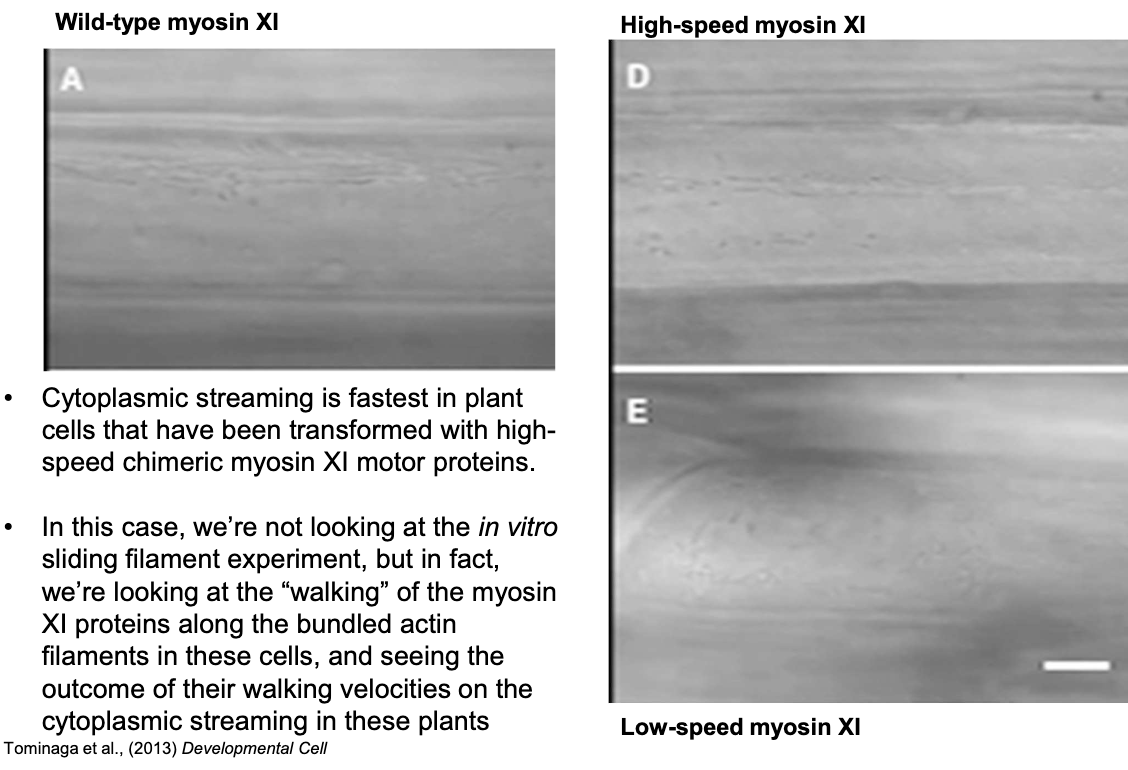
What role does cytoplasmic streaming play in plant cell size and development?
It regulates cell size by enabling efficient material transport
Especially important in large aquatic plant cells (Elodea and Chara) that have limited diffusion ability
Slower in land plants (larger number of small cells)