L29 Microbial Population Growth
1/21
Earn XP
Description and Tags
Vocabulary flashcards covering key terms from the lecture on microbial population growth, including binary fission, growth phases, nutritional requirements, and methods of study.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
22 Terms
Binary Fission (the prokaryotic “mitosis”)
Asexual reproduction by cell division in prokaryotes, resulting in two genetically identical cells.
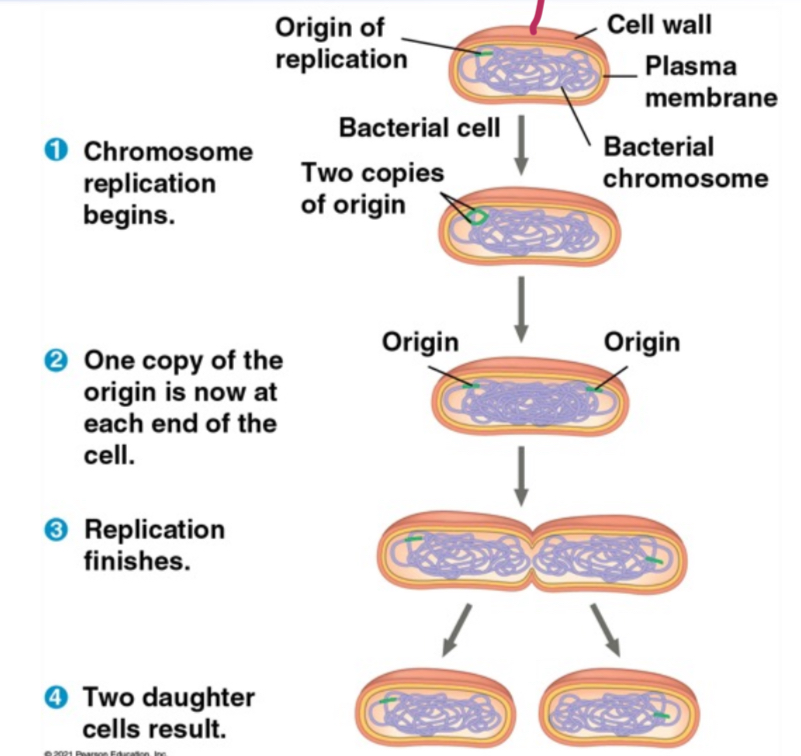
Why prokaryotes are so dominant
Smaller genome= faster replication (13 minute doubling time) so evolve/adapt fast eg. antimicrobial resistence. Fast growth+ 3.5 billion years= colonization of all ecosystems.
Microbial growth
Not much different from eukaryotes in cellular requirements. Microbes need the same building blocks, just in different amounts, when supplied with all required materials they can reproduce via binary fission.
Closed Batch Culture System “classic” microbial growth
Standard method of studying microorganisms in culture with a defined, limited supply of nutrients. Once used, cells cannot proliferate. Bias towards fast growing organisms eg paothogens.
Lag Phase
Initial phase in a batch culture where cells require time to initiate biosynthetic reactions before active division. Length depends on history of inoculum.
Exponential (Log) Phase
Cells are actively dividing with no growth limitations. Population doubles in a constant time interval under ideal conditions.
Stationary Phase
Cells stop growing; cryptic growth is observed with an equilibrium between growing and dying cells (dynamic population).
Cryptic growth
Organisms survive by consuming lysed cell constituents of other dead cells within the culture.
Death Phase
Cell death occurs, and equilibrium is skewed towards death.
Carbon Source (material to build with)
Building blocks for macromolecular synthesis required for microbial growth.
Energy Source (to build)
Energy (electrons) to drive anabolic and catabolic reactions in the cell required for microbial growth.
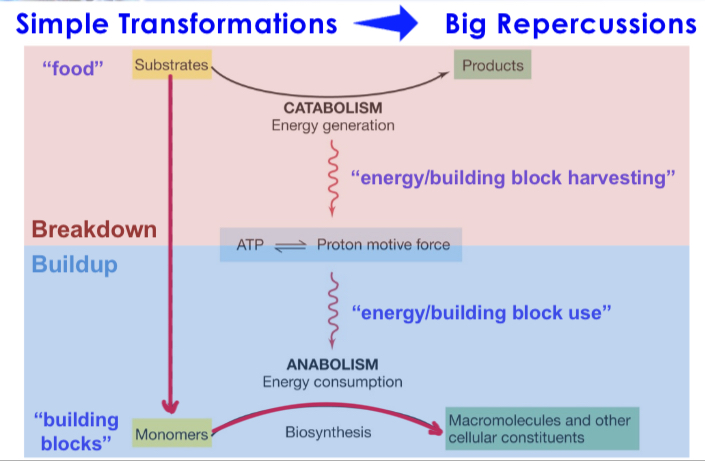
Reducing Power
Carriers of energy/electrons (NAD+/NADP+) required for microbial growth. (NADH=reduced form)
source of carbon and energy
Light reaction, light energy transformed into NADPH and ATP. Harvest carbs from autotrophs as enery/carbon sources. Redox reactions.
Redox reactions
Chemical energy stored in bonds, when broken can be captured in new bonds (ATP), can be broken for later use. This (re)duction and (ox)idisation of coupled compounds can be applied to many compounds.
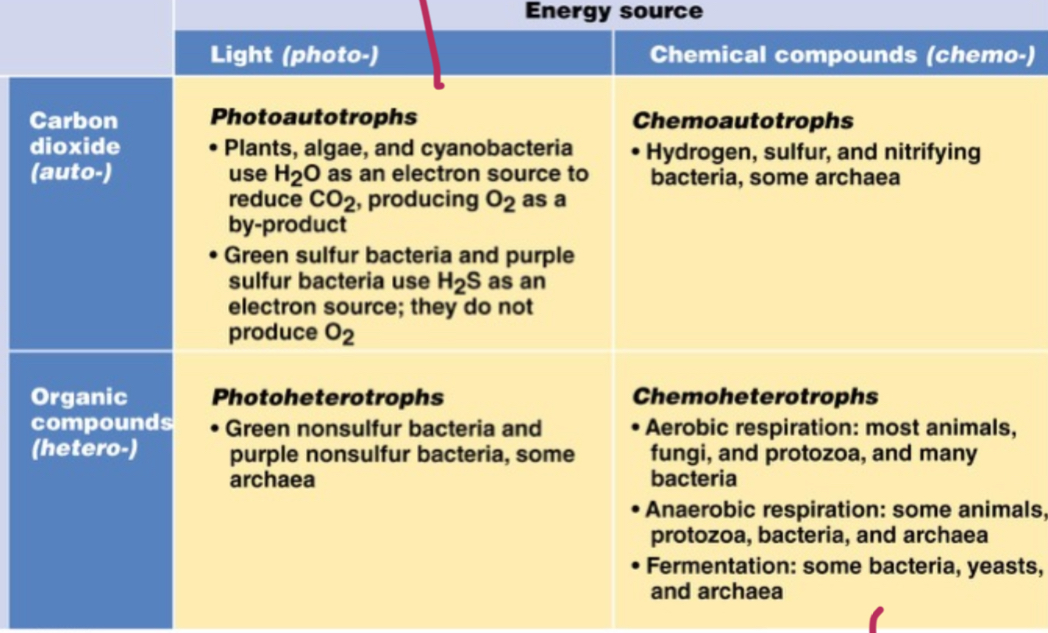
Trophic (nourishment) groups in microbiology
Carbon source; carbon dioxide= (auto), organic compound- (hetero). Energy source; light= (photo), Chemical compounds= (chemo). Add troph to the end.
Auxotroph
An organism that is unable to synthesize one or more essential growth factors, and it will not grow unless limiting factor is provided
Wild type strain
Has all essential genes, can grow by itself, can be isolated into pure culture.
Cross-feeding (Syntrophy)
When one species gains metabolic products of another species, allowing for survival of auxotrophs. Interactions can benifit one or both species. Common in nature and partially explains inability to culture most microbes.
Microbiome
The complete collection of microorganisms, and their genes, within a particular environment.
Microbiota
Individual microbial species in a biome – bacteria, fungi, archaea and viruses.
Culture-dependent methods (using pure cultures)
Relies on culturing of microbes in the lab. Pro: Can study one organism at a time, allows access to phenotype, can manipulate environment to see response. Con: Not all organisms can be cultured, too many species to grow them all, inconsistent with real world conditions, requires precise conditions to match microbiome needs.
Culture-independent methods (uses sequencing or metabolic profiling)
Relies predominantly on nucleic acid-based methods. No culturing required. Pro: Can study many organisms at a time, allows acces to genotype, shows communities as they are in nature, can target non-culturable organisms, provides access to unknown info/species. Con: No pure culture so no ability to manipulate, expensive and complex methods.