Lecture 4 (Chapter 16): Regulation of gene expression
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
Key concepts
prokaryotic gene expression is regulated in operons
eukaryotic gene expression is regulated by transcription factors
epigenetic changes regulate gene expression
eukaryotic gene expression can be regulated after transcription
Two ways to regulate a metabolic pathway
Allosteric regulation
Regulation of protein synthesis
Allosteric regulation
regulation of enzyme-catalyzed reaction (enzyme activity) allows rapid fine-tuning
Regulation of protein synthesis
slower regulation but conserves energy and resources since protein synthesis requires a log of energy (transcription regulation via regulation of enzyme concentration)
Alternative method for controlling an enzyme/cell function
acetylation, methylation, phosphorylation
Where is prokaryotic gene expression regulatied?
in operons
Do all promoters have the same recognition sequences?
No! different classes of promoters have different recognition sequences (ex. -35 and -10)
What must happen before transcription can begin?
DNA sequence binds RNA pol and sigma factors to enhance RNA pol binding at certain promoters
RNA pol by be bound to sigma factor before it can recognize a promoter and begin transcription
What is Sigma-70?
a sigma factor that is active most of the time and bins to recognition sequences of housekeeping genes (genes normally expressed in actively growing cells)
What is an example of an enzyme that can induce the expression of specific gene(s)?
lactose! In e. coli the intestine must adjust quickly to changes in food supply. When lactose is added, mRNA for the enzyme is made, once it’s removed, the amount of mRNA is decreased
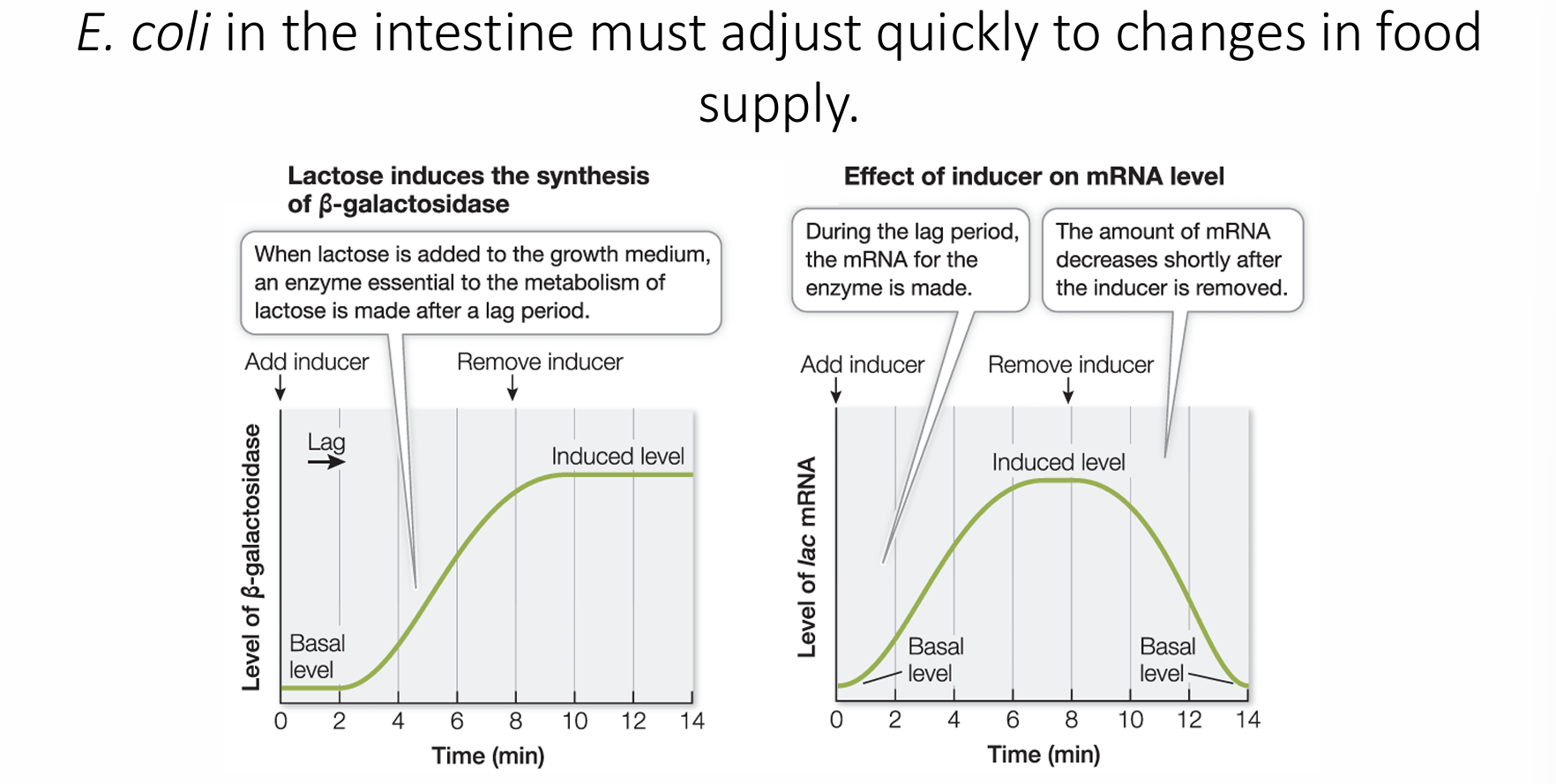
Two types of prokaryotic gene regulation
inducible systems
repressible systems
inducible systems
substrate (inducer) interacts with a regulatory protein (repressor); repressor can’t bin to operator and transcription proceeds
repressible systems
a product (co-repressor) binds to a regulatory protein, which then binds to the operator and blocks transcription
What is regulated gene expression in prokaryotes?
prokaryotes made some proteins only when they are needed through transcription regulation. regulatory proteins bind to promoters
Negative regulation
a repressor protein prevents transcription (without, doing transcription)
positive regulation
an activator protein stimulates transcription (without, not doing transcription)
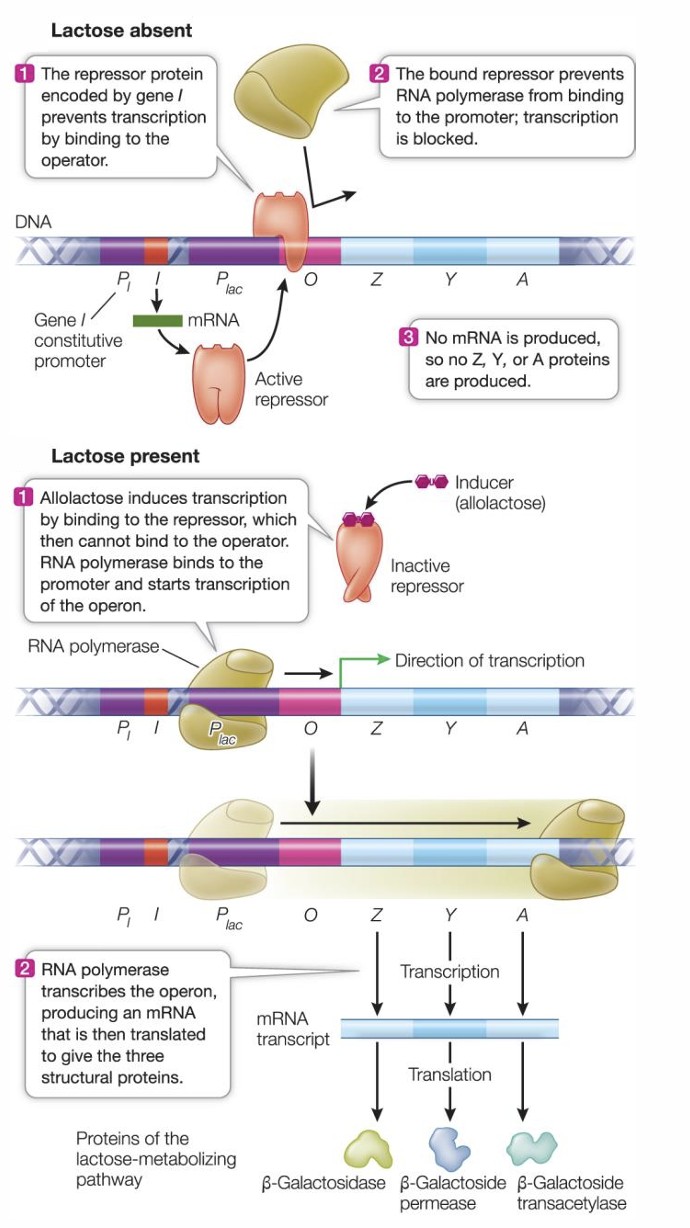
What type of system is the lac operon?
it is an inducible system (can be turned on)
Operon
gene cluster with a single promoter
What does an operon consist of?
a promoter
two or more structural genes
an operator (short DNA sequence between promoter and structural genes that binds to regulatory proteins)
Structural genes
specify a primary protein structure (the amino acid sequence). Three structural genes for lactose enzyme are adjacent to the chromosome and share a promoter
Lac operon
the repressor has two binding sites, one for the operator (actual DNA sequence in operon), and one for the inducer (allolactose; small molecule that binds to repressor)
When lactose is absent, the repressor prevents binding of RNA polymerase to the promoter which blocks transcription
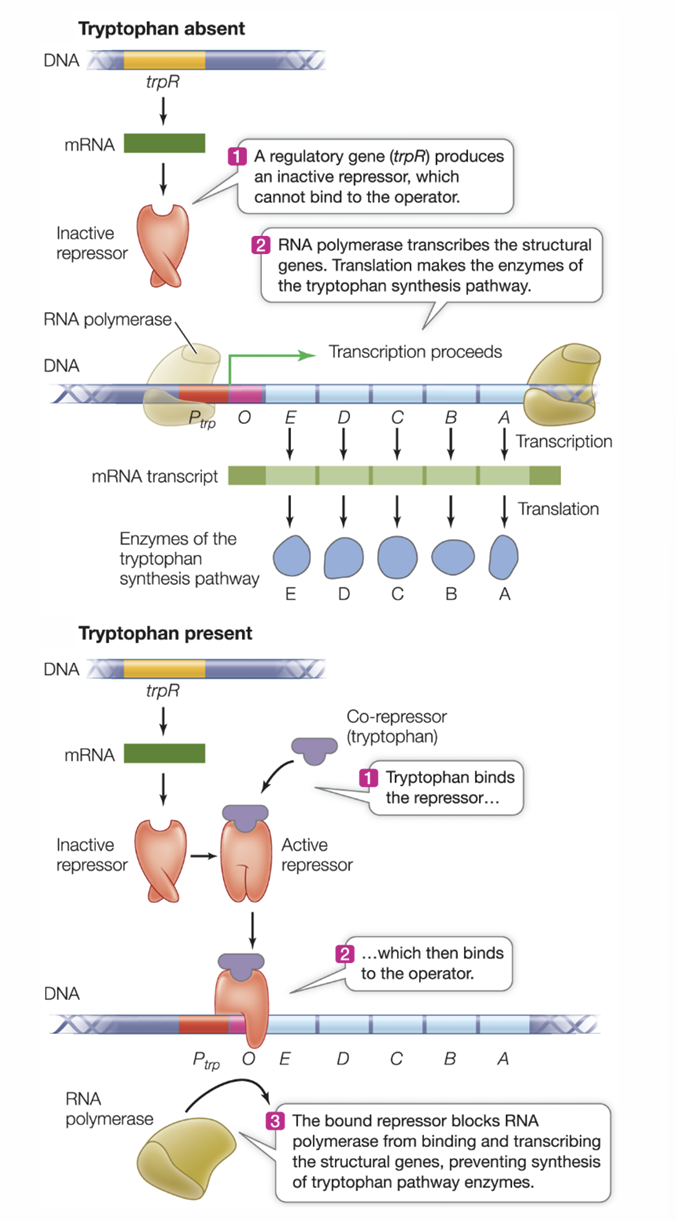
trp operon
a repressible system that is always on unless repressed
the genes code for enzymes that catalyze synthesis of tryptophan
when there is enough tryptophan in the cell, tryptophan binds to the repressor which then binds to the operator
tryptophan is a co-repressor (helps shut down gene expression but doesn’t bind to operon directly)
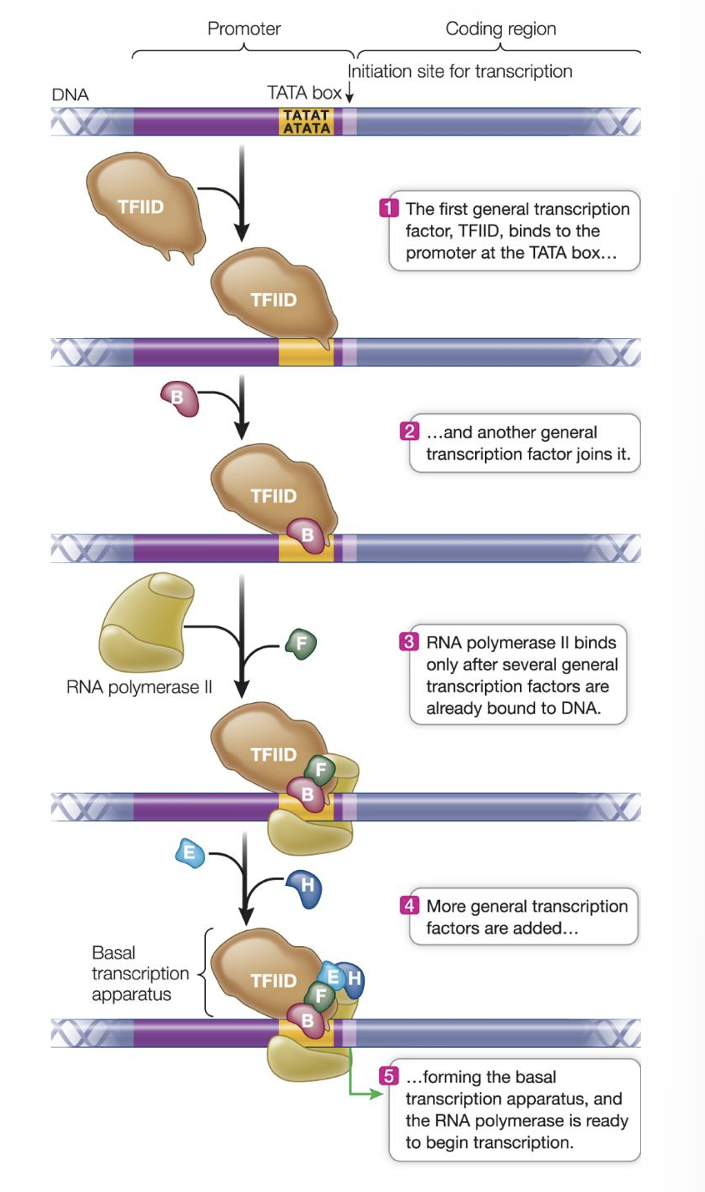
What regulates eukaryotic gene expression?
transcription factors! expression of eukaryotic genes must be precisely regulated, which can occur at several different points
Eukaryote promoters
sequences near the 5’ end of the coding region, many have a sequence called the TATA box where the DNA begins to denature. promoters also include regulatory sequences recognized by transcription factors
Regulation of RNA polymerase II binding
RNA pol II can only bind to the promoter after general transcription factors have assembled on the chromosome
TFIID binds to the TATA box first, then the other factors bind to form an initiation complex
each general transcription factor has a role in gene expression
Enhancers
regulatory sequences that bind transcription factors that activate or increase the rate of transcription (positive regulation)
Silencers
bind transcription factors that repress transcription (negative regulation)
Where are regulatory sequences located in eukaryotes?
thousands of base pairs away, and may affect the expression of several nearby genes
mediator
a protein that binds to the basal transcription apparatus and causes DNA to bend, bringing regulatory sequences close to the promoter
Requirements of structural motif for DNA recognition
must fit into a major or minor groove
have amino acids that can project into the interior of the double helix
have amino acids that can form hydrogen bonds with interior bases
structural motifs are a recurring protein shape that enables DNA binding or protein interaction
What mediates cell differentiation?
transcription factors that determine gene expression
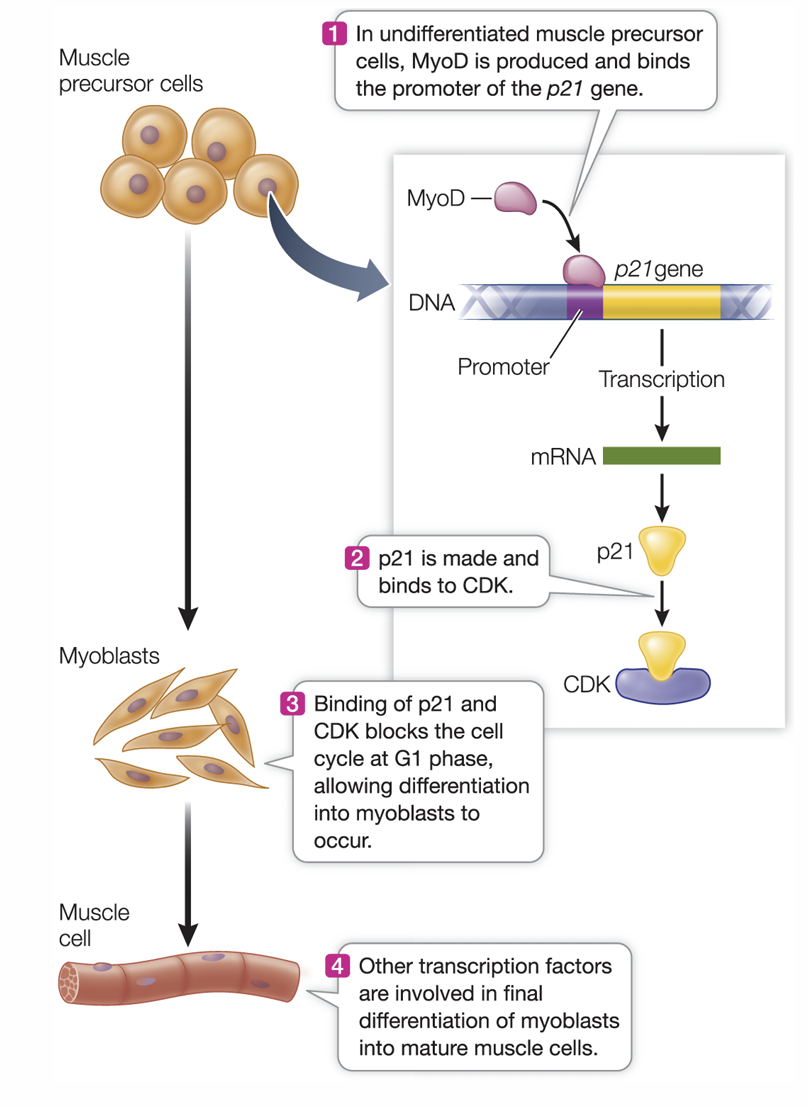
Formation of muscle cells
transcription factors fist inhibit cell division, the first step in differentiation
other transcription factors control the other processes of differentiation
How do eukaryotes coordinate expression of sets of genes?
most have their own promoters, and the genes may be far apart in the genome
if the genes have common regulatory sequences, they can be regulated by the same transcription factors
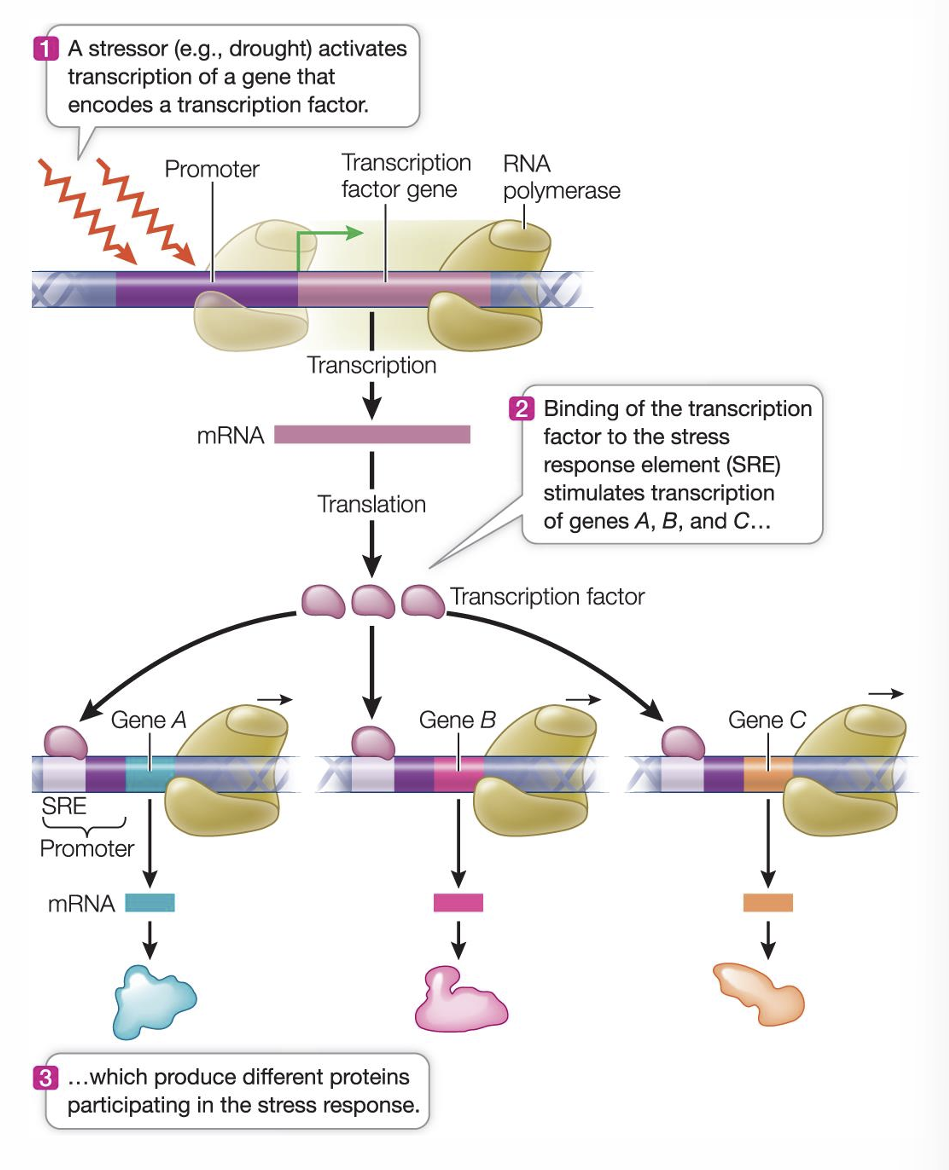
Example of coordinated gene expression in plants
plants in drought stress must make several proteins which genes are scattered throughout the genome
each of the genes has a regulatory sequence called stress response element (SRE)
a transcription factor binds to the SRE and this element stimulates mRNA synthesis