Biochem 285 Week 7
1/19
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
20 Terms
What are DNA damage and mutation most frequently caused by?
Mismatch base pairing that is not resolved by DNAP, spontaneous mutations (UV light or chemical reactions), and double stranded breaks (UV light, enzymatic cleavage, breakage during replication).
Mismatch Repair Pathway (MMR)
Evolved to fix the problem of DNAP missing an incorrect base addition to avoid the change from being permanent. It becomes active after DNA replication.
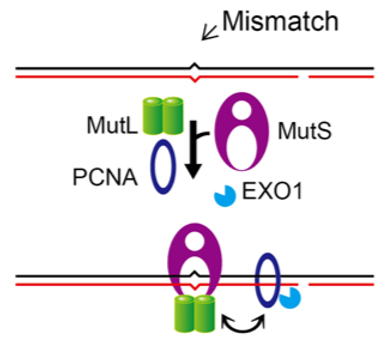
MMR in Prokaryotes
Mis-matches in DNA result in kinks. Kinks are then recognized by MutS and then MutS phones its friend MutL to come and look. Methylase enzymes then come in and add methyl groups to the older, template strand after DNA replication. Then MutH comes in and recognizes the methylation pattern on the older strand. MutH however, needs help as an endonuclease and so it interacts with MutL to become activated and then it cuts the new strand of DNA. Lastly, a helicase, UvrD, helps unwind the DNA and an exonuclease, Exol, removes the bases just beyond the mismatch. DNAP and ligase then complete the repair.
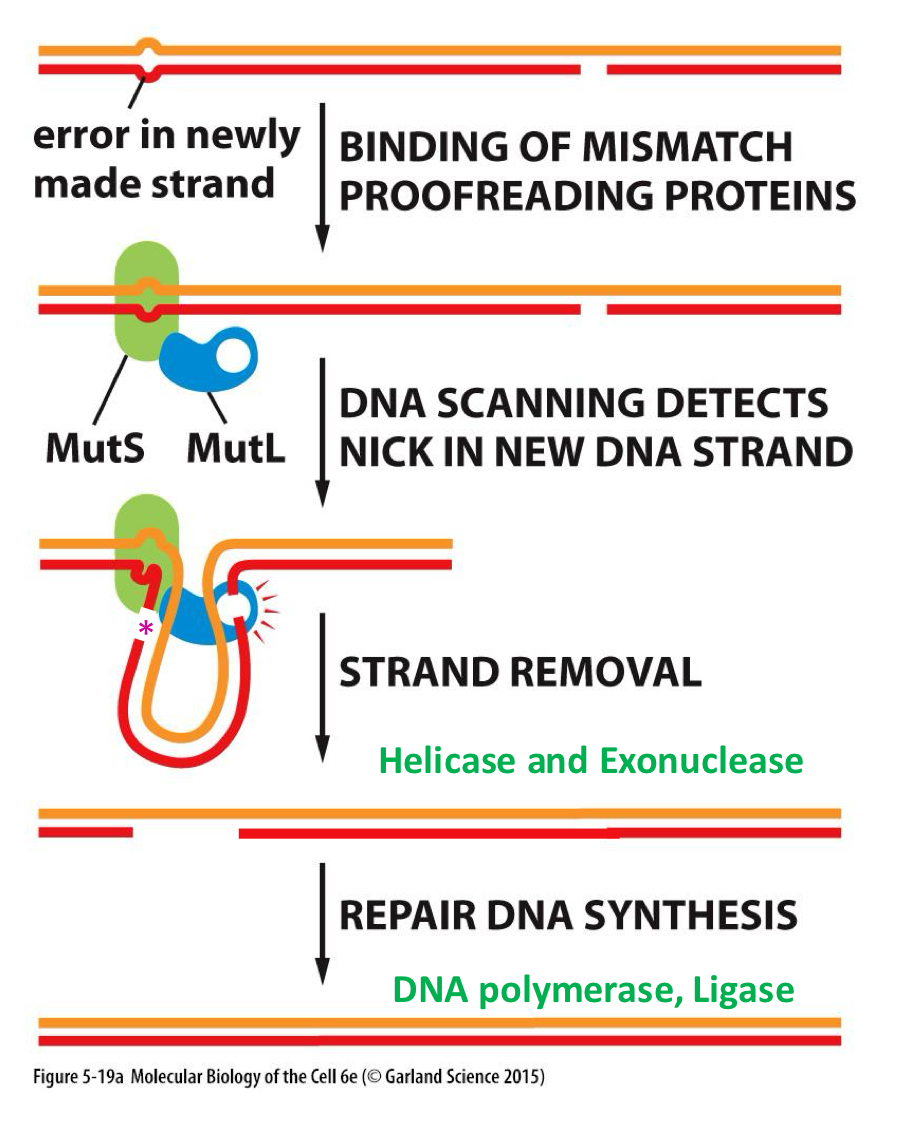
A mnemonic to remember the order of proteins in MMR (prokaryotic)
Stop (MutS), Letting (MutL), Hungry (MutH), Dogs (UvrD), Exit (Exol)

Depurination
A type of spontaneous DNA damage where a purine base is lost from a DNA molecule. Most likely to occur on promoter region. Note: This only happens to Adenine and Guanine because there are purines.
Deamination
A type of spontaneous DNA damage where an amine group (-NH2) is lost from a DNA molecule. This happening in C, A, or G leads to unnatural DNA bases. Most likely to occur on promoter region. Cytosine to Uracil, Adenine to Hypoxanthine, and Guanine to Xanthine. Thymine cannot be deaminated as is has no amine group.
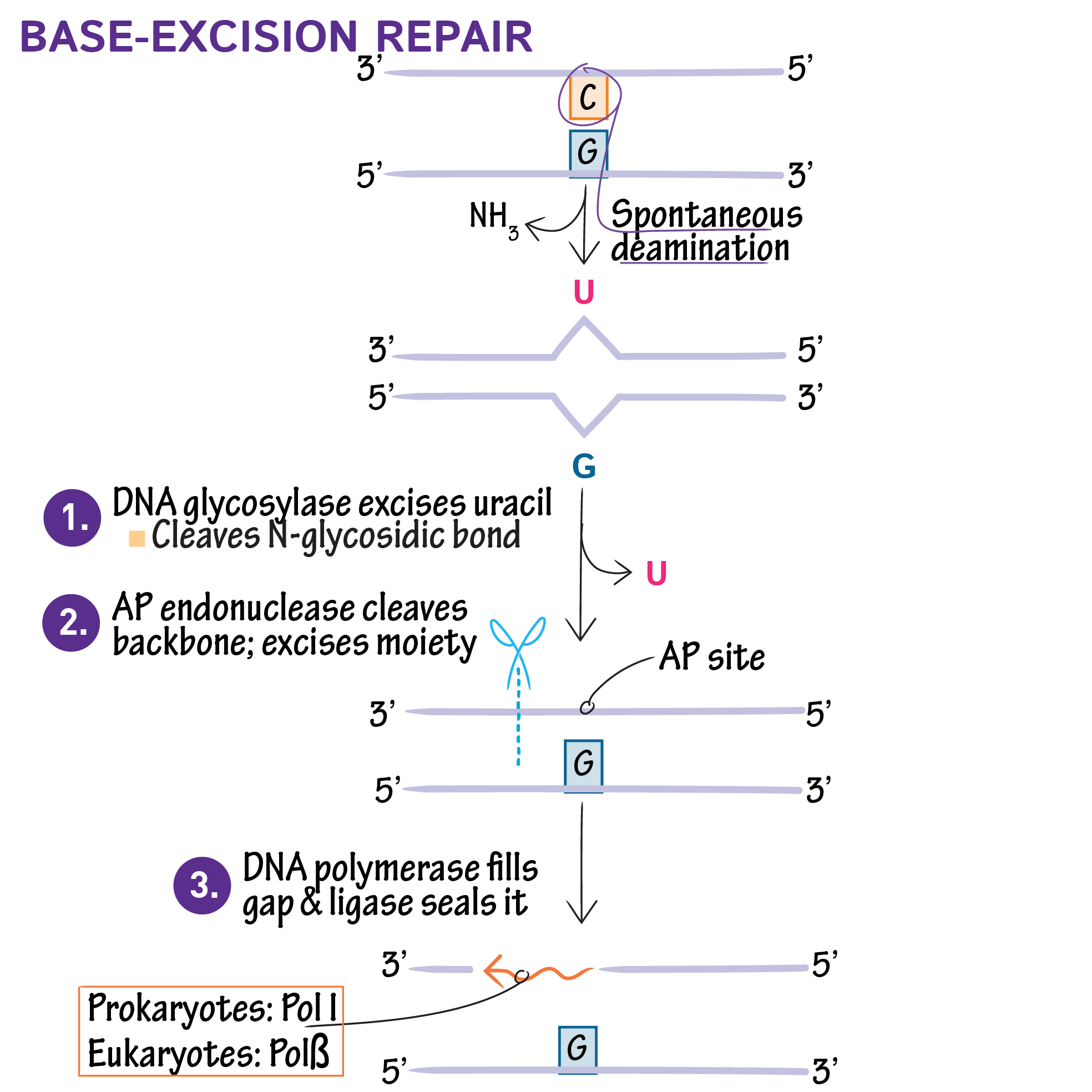
Base Excision Repair (BER)
It is used to replace one or few damaged DNA bases that happened through depurination or deamination. DNA glycosylase will recognize the damaged base (on either the new or old strand) and flip it out of the strand and then cleave the glycosyl bond that connects the base to the backbone. After the base is removed, and AP endonuclease will remove the remaining sugar phosphate backbone. DNAP will then come in and add the correct base, ligase will seal the nick.
What type of bond connects the base to the ribose backbone is a DNA molecule? What enzyme will break it?
Glycosyl bond; DNA Glycosylase
What type of bond connects the phosphate group the ribose in a DNA molecule? What enzyme will break it? What enzyme will fix/create it? (Hint: this bond makes the DNA backbone)
Phosphodiester bond; Nuclease; Ligase
What type of bond connects one base to another base in a DNA strand? What enzyme will break it?
Hydrogen bond; DNA Helicase
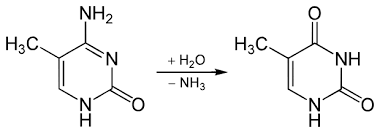
What does deamination of 5’ methyl-cytosine create?
Thymine. This mutation leads to a T:G mismatch.
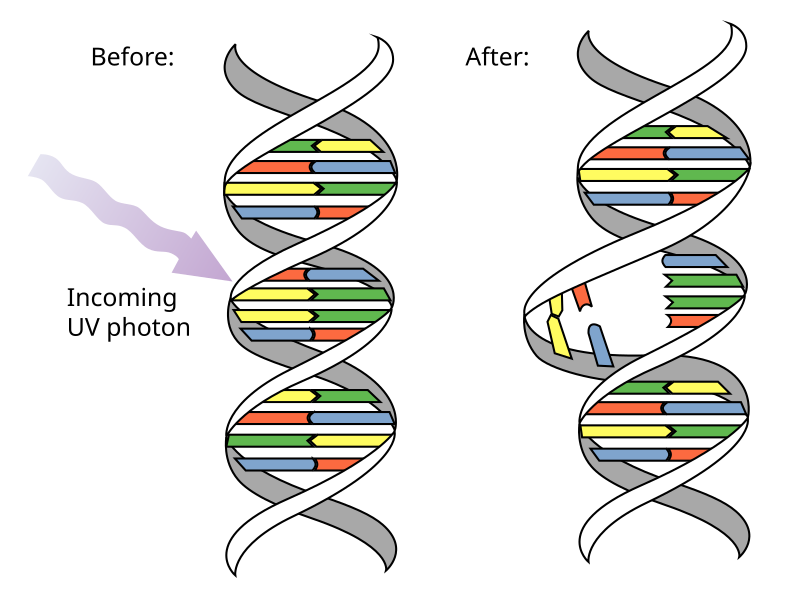
UV Radiation
A type of spontaneous DNA damage where radiation can cause covalent bonds to form between pyrimidines (pyrimidine dimers). Pyrimidine dimers distort DNA structure, cause RNAPs and DNAPs to stall, and replication and transcription are either inhibited or errored.
Nucleotide Excision Repair (NER)
Fixes the occurrence of pyrimidine dimers in DNA strand after radiation damage.
Xeroderma Pigmentosum
An autosomal recessive disorder when there is mutations in the genes needed for the NER pathway. Increase in skin lesions and likelihood of skin cancer.
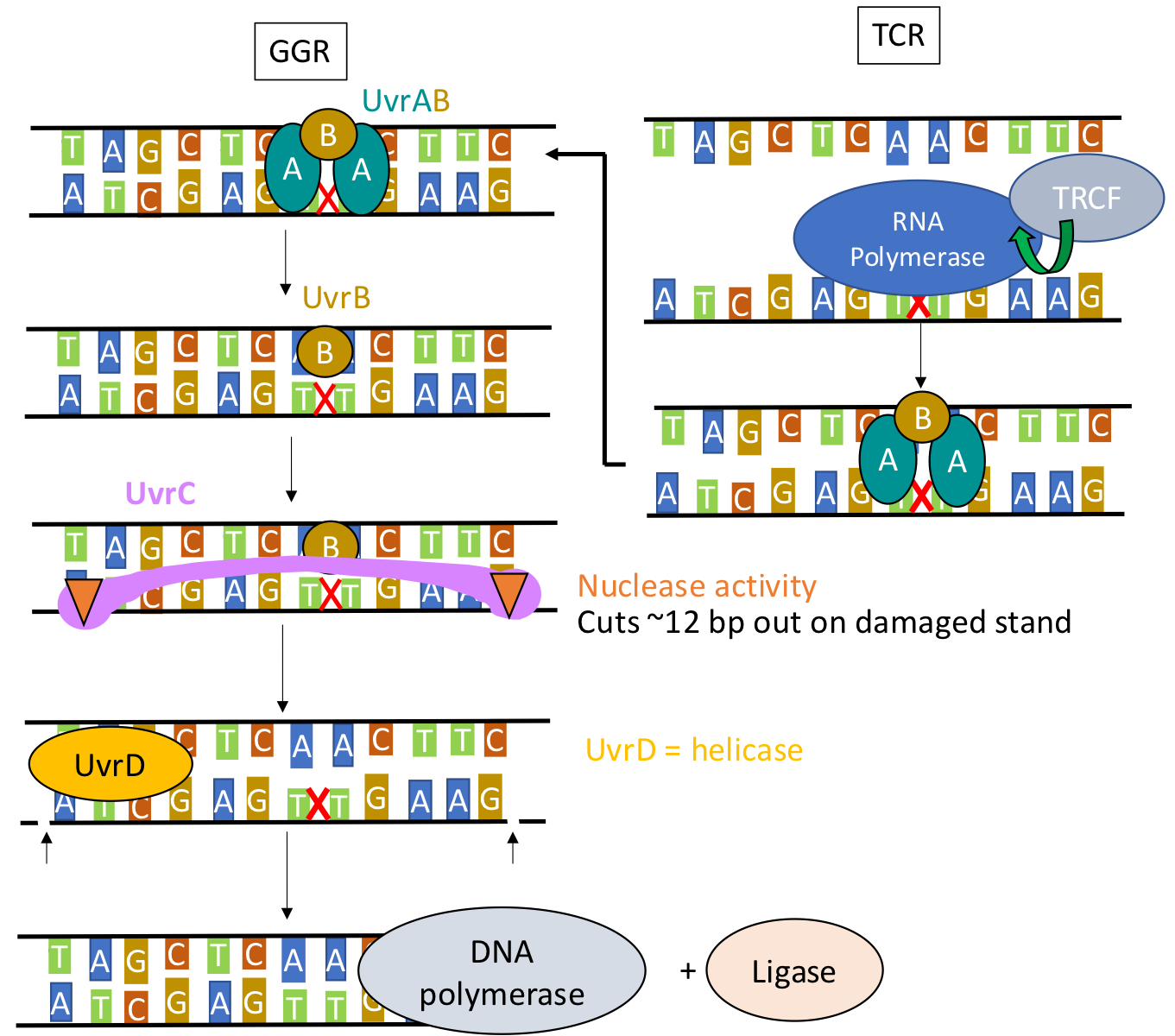
NER in prokaryotes (E. coli)
One of two ways will be used to identify pyrimidine dimers at first: global genome repair or transcription coupled repair. Transcriptional repair coupling factor then recruits UvrAB proteins to the site. UvrA leaves and UvrB recruites UvrC. UvrC is a nuclease and will cut out ~12 bps on damaged strand. UvrD is a helicase and will unwind the cutout. Lastly DNAP and ligase will come in to finish the repair.
Double-stranded DNA break
A type of spontaneous DNA damage where both DNA strands in a helix are broken. Occurs frequently during replication.

Non-Homologous End-Joining (NHEJ)
The predominant repair mechanism for double-stranded DNA breaks in eukaryotes. The protein Ku recognizes broken DNA ends through interactions with the sugar-phosphate backbone and its own heterodimeric structure. Then a nuclease trims back base (losing some DNA) and a ligase connects the backbone back.
Homology Directed Repair (HDR)
Repair pathway that utilizes a homologous template (one that has a similar sequence and structure to the broken DNA region) and makes a more seamless repair, therefore leading to less DNA sequence loss. The homologous template is from a sister chromatid or a homologous chromosome. This pathway is also most active during replication (S phase) or just after replication (G2 phase) because of the templates available (this is a potential limitation).
Steps of HDR
(MRN and CtlP) Nuclease complexes chew back the broken DNA strands and RPA binds to the ends. Then, RAD51 binds and the single strand invades a homologous DNA strand. It base pairs and DNAP comes in and begins to copy. When the two new strands are long enough, they separate from the sister chromatid and base pair with each other serving as primers for DNAP. Lastly, the nicks are sealed by ligase.
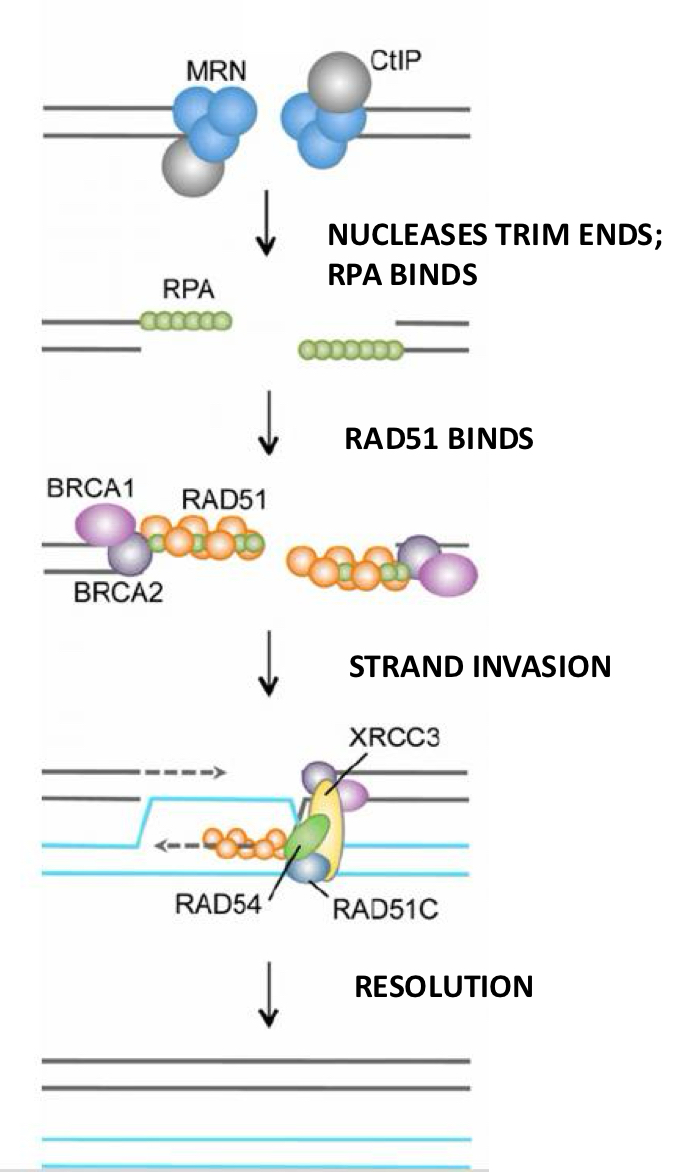
BRCA2
a scaffold protein used in HDR that helps to bring RAD51 rapidly to sites of damage and release the active RAD51 onto single strand DNA when necessary
