MCB*2050 #2 - Activators, repressors, chromatin, and epigenetics
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
27 Terms
How is the expression of eukaryotic genes regulated?
Through multiple protein-binding transcription-control regions located at various distances from the transcription start site
These control regions can be close to the transcription initiation site (called promoter-proximal elements) or distal sites (called enhancers)
promoter-proximal elements and enhancers are gene and cell-type-specific regulatory elements on DNA
Transcription activators and repressors are are modular proteins containing a DNA-binding domain and one or more activation or repression domains
How do we find the gene regulatory sequences?
linker scanning-reporter gene technique for identifying gene regulatory elements on DNA
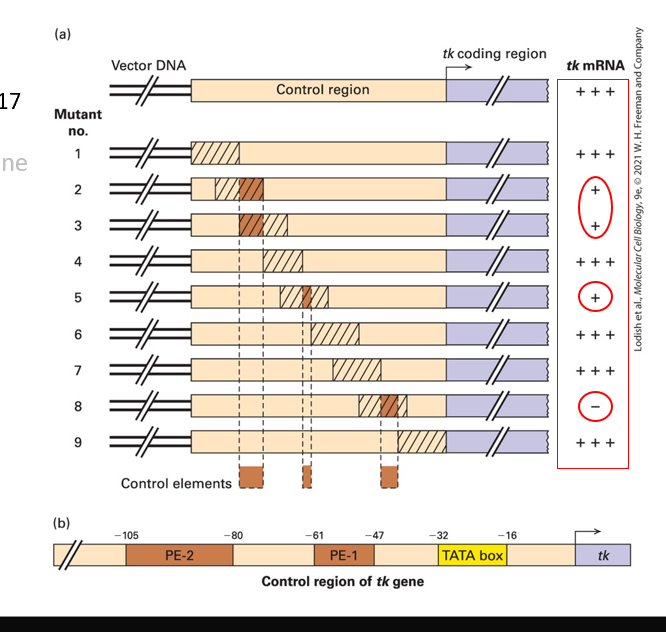
What do these tests indicate?
Promoter-proximal elements regulate eukaryotic gene expression.
Eukaryotic promoter region DNA that supports high-level expression of a reporter gene (light purple) is cloned in a plasmid vector
overlapping linker scanning (LS) mutations (insertions of scrambled sequence) are introduced from one end of the region to the other
mutant plasmids are transfected separately into cultured cells
The expression/activity of the reporter gene product
The results are…
•LS mutations 1, 4, 6, 7, and 9 have little or no effect on expression of the reporter gene, indicating that regions altered in these mutants contain no control elements.
•In LS mutations 2, 3, 5, and 8, reporter-gene expression is significantly reduced, indicating that the regions contain control elements.

Describe the general organization of control elements that regulate gene expression in multicellular eukaryotes and yeast
•Most eukaryotic genes are regulated by multiple transcription-control elements.
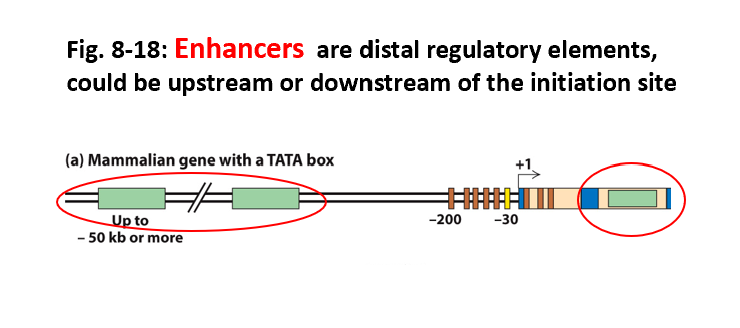
•(a) Mammalian genes with a TATA box promoter:
•Regulated by promoter-proximal elements and enhancers.
Promoter elements: → Position RNA polymerase II to initiate transcription at the start site.
Influence the rate of transcription.
Enhancers may be either upstream or downstream (or in introns) and as far away as hundreds of kilobases from the transcription start site.
•Promoter-proximal elements are found upstream and downstream of transcription start sites at equal frequency in mammalian genes.
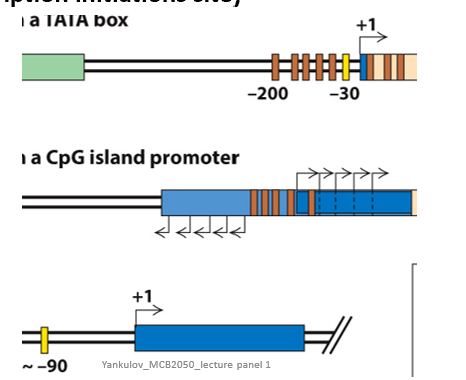
•(b) Mammalian genes with a CpG island promoter:
•Transcription initiates at several sites in both the sense and antisense directions from the ends of the CpG-rich region.
•Sense direction transcripts are elongated and processed into mRNAs by RNA splicing.
•Genes express mRNAs with alternative 5′ exons determined by the transcription start site.
•Promoters contain promoter-proximal control elements. (Not clear whether they are also regulated by distant enhancers.)
•(c) Most S. cerevisiae genes:
•Contain only one upstream activating sequence (UAS) and a TATA box, ~90 bp upstream from the transcription start site.
Describe the location of promoter-proximal regulatory elements
Close to the transcription initiation site
Describe the location of enhancers
distal regulatory elements, could be upstream or downstream of the initiation site
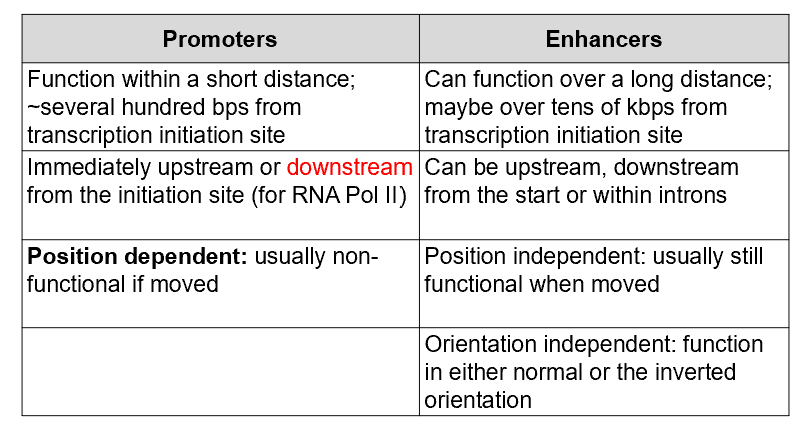
What are the properties of promoters and enhancers?
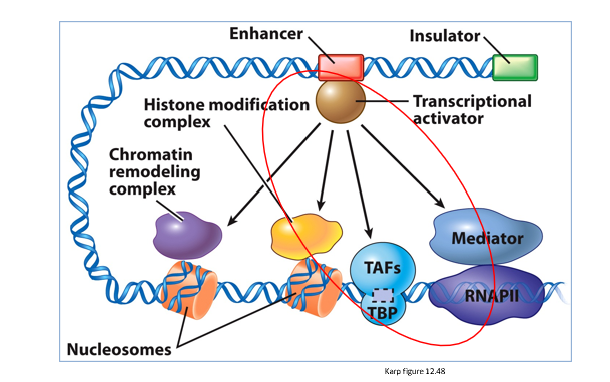
How do enhancers communicate with promotors?
communicate via bending DNA
True or False: transcriptional activators and repressors are proteins with distinct domains
True!
Transcriptional activators or repressors often have independent DNA-binding domains and effector (activation or repression) domains
These can be identified by deletion analysis of the protein
What is a domain?
a part of a protein that will aquire a distinct 3D structure and distinct activity within the whole protein
portion of a polypeptide with a specified function
identified by deletion analysis of actual peptides
What is a motif?
A sequence in a protein that, based on similarity with other known functional domains, is likely to produce a distinct 3D structure and a distinct domain
the majority of DNA binding domains and activation domains have conserved motifs
a conserved peptide sequence found in DOMAINs with similar functions
Describe the modular structure of eukaryotic transcription activators
Transcription factors may contain more than one activation domain but rarely contain more than one DNA-binding domain:
Gal4 and Gcn4 are yeast transcription activators.
Glucocorticoid receptor (GR) promotes transcription of target genes when glucocorticoid hormones bind to the C-terminal activation domain.
SP1 binds to CpG promoter elements in a large number of mammalian genes.
What is a Zn++ finger motif
→ regularly spaced C and H amino acids
(histidine and cysteine)
forms a domain for binding to DNA
What is the Leu zipper motif?
→ found in protein dimerization domains: regularly spaced L amino acids residues
(Leucine)
forms a domain for dimerizing with another protein and then to DNA
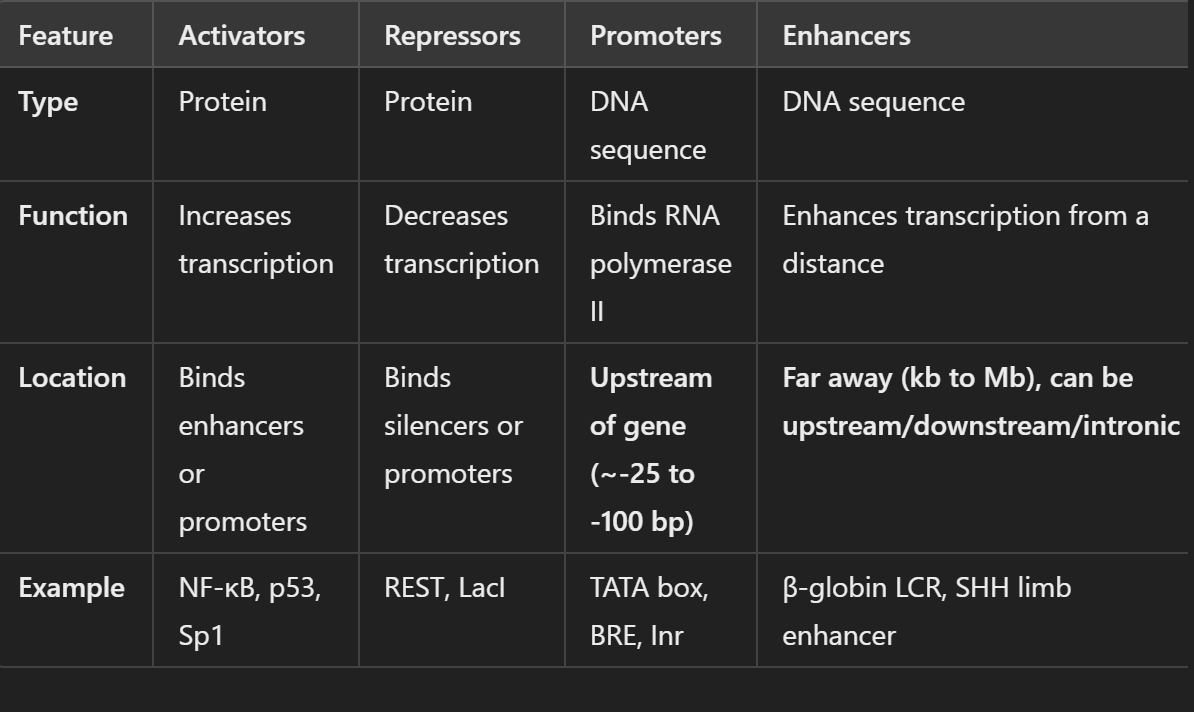
Describe the difference between enhancers, repressors, promoters, and activators
they’re basically different in how they affect gene expression

Describe the combinatorial binding to DNA of different dimer combinations of proteins
•Very often DNA-binding proteins of the same class (for example Helix-Turn-Helix) work as dimers.
•Each monomer (a single molecule that can bind to other molecules to create a polycule) can recognize a specific DNA element, but cannot work on its own. It will bind to this DNA only in a complex with a similar protein.
•This means that if two proteins (green and yellow) can combine, they will have three binding sites (gg, yy, gy), three proteins (green, yellow, blue) can have six binding sites (gy, gb, yb, gg, yy, bb), etc…
dimerization of a similar protein increases the number of DNA-binding options in this group of proteins
True or false: if you have three monomers, you can form dimers
True!! you an have six combinations of these proteins
•These six combinations can recognize six DNA binding sites
•Adding an inhibitor for one of the three proteins increase the possible outcomes for activation (sort of a ‘fine tuning’)
What are enhanceosomes?
combinatorial binding to DNA-binding proteins from different classes
eukaryotes have very complex promoters and enhancers
in these, various transcription activators and repressors often interact with each other and cooperate to establish a strong activator or repressor site to tightly control the expression of the target genes
overall, it is a multi-protein complex that assembles on an enhancer to regulate gene transcription. It acts as a highly organized structure where transcription factors (TFs) work together to activate transcription efficiently.
What is the general design of transcription factors in the nuclear-receptor superfamily?
They have a DNA binding domain and a ligand binding domain
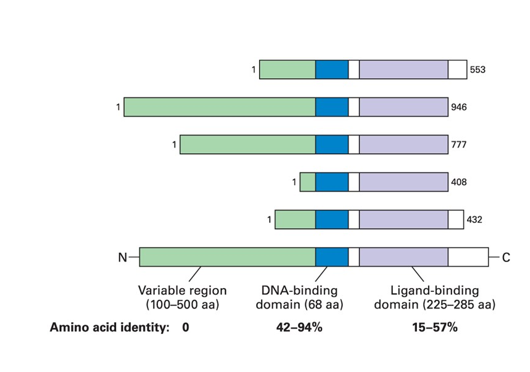
Describe how transcription factor activity can be directly regulated by lipid hormones
•A cell sends a signal to other cells by secreting a lipid soluble hormone
•Lipid-soluble hormones diffuse into the cell through the cell membrane and bind to a dedicated class of proteins: the Lipid-Hormone Receptors/ Transcription Activators
•Some receptors are located in the cytoplasm
oUpon binding of the hormone the receptors-hormone complex moves to the nucleus. It is now a transcription factor - it binds to specific response elements in genes to regulate gene expression.
•Other receptors are located in the nucleus, the binding of the hormone changes their activity
→ the hormone-binding domain is sufficient to translocate the protein to the nucleus
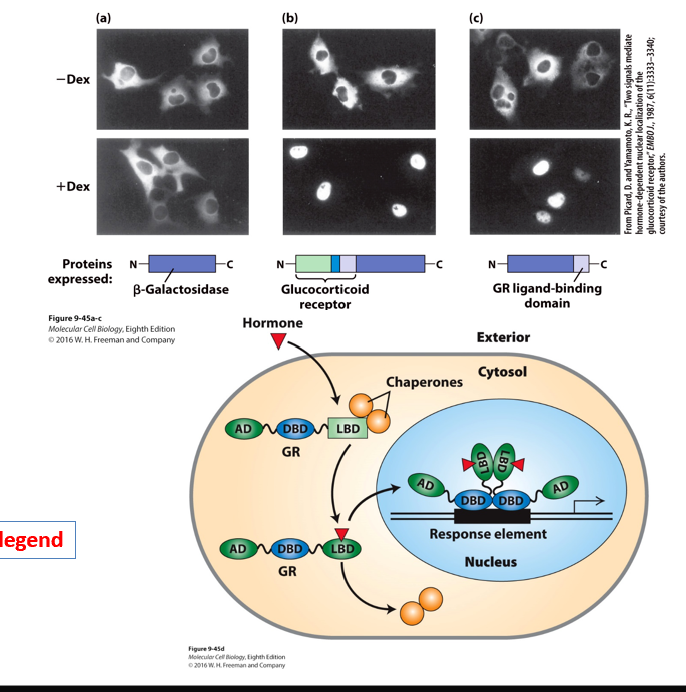
What is this diagram discribing?
Fusion proteins demonstrate that the hormone-binding domain of the glucocorticoid receptor mediates translocation to the nucleus in the presence of hormone.
•Heterodimeric nuclear receptors , Located exclusively in the nucleus.
Where the hormones enter/leave…
•(–) Hormone – repress transcription when bound to their cognate sites in DNA by directing histone deacetylation at nearby nucleosomes
•(+) Hormone – conformational change – binds histone acetylase complexes – reverses repressing effects
•Homodimeric receptors:
•(–) Hormone – in the cytoplasm
•(+) Hormone – receptors translocate to the nucleus and bind to response elements
glucocorticod receptor (GR) translocates to the nucleus in the presence of hormone
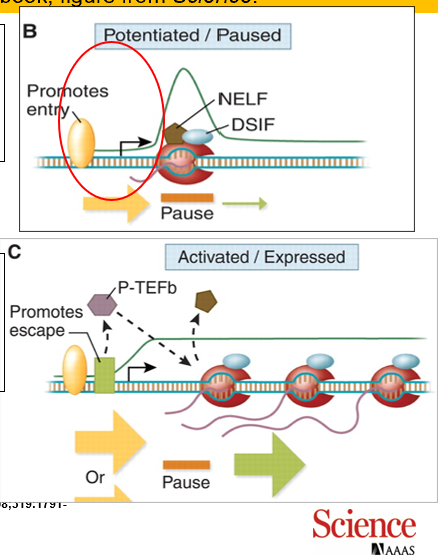
What does this diagram demonstrate?
The first photo discribes how RNA polymerase elongation is blocked, awaiting a signal by a transcriptional activator
→the second photo demonstrates how activators stimulate PTEFb, and how RNA pol II block is released and the gene is once again active
Overall, this is demonstrating the PAUSE that occurs when RNA pol II initiates transcription and then pauses
How does eukaryotic activators/repressors usually bind directly to DNA in promoters or enhancers?
they affect gene expression by recruiting multi-subunit co-activator/co-repressor complexes that modulate chromatin structure or interact with the Mediator and the General Transcription Factors.
activators stimulate the assembly of preinitiation complexes and move nucleosomes away from the promoters
a cell needs to produce the specific set of activators required for a specific promoter/enhancer of a particular gene to express that gene
Repressors most frequently work through the build-up of repressive chromatin structures
Describe quickly once again, what Euchromatin and heterochromatin is
Heterochromatin:
regions of chromosomes that are more intensely stained
DNA is more densely packed and it is rich in repetitive DNA mostly located in centromeres and telomeres
not accessible to transcriptional machinery
genetically inactive
Euchromatin:
lightly stained chromosome regions
where most eukaryotic genes are locted
genetically ACTIVE
What central role does chromatin play?
in the selection of genes that will be expressed in any tissue or cell
in the activation or repression of genes by DNA binding proteins
Recall epigenetic regulation and pioneer transcription factors
epigenetic regulation:
Transcription in eukaryotes takes place in chromatin, chromatin needs to open for transcription to proceed
may interact with multiprotein co-repressor complexes to condense chromatin
Pioneer transcription factors:
establish and maintain open chromatin structure. They do not directly activate transcription
binds to a specific regulatory sequence within the condensed chromatin
Interacts with chromatin-remodeling enzymes and histone acetylases that decondense the chromatin, making it accessible to RNA polymerase II and general transcription factors.

How many histones is the nucleosome built of? and describe them
8 histones
two of each - H2A, H2B, H3 and H4
•The histone N-termini are not structured and protrude away from the histone core. These N-termini are modified by post-translational modifications (PTMs).
The PTMs dictate the function of the DNA wrapped around the nucleosome.
•The variety of PTMs are referred to as “histone codes”.