MCB 250 – RNA Interference
1/29
Earn XP
Description and Tags
Module 37: How was RNAi discovered? (Nobel prize in 2006) • How is using RNAi experimentally different from making a knockout or generating mutants? • What does Dicer do to foreign dsRNA? • What does the RISC complex/Argonaut do? • What do Drosha and Dicer do to pri-miRNAs and pre-miRNAs, and how do they recognize their targets? • What is the distinction among siRNAs, miRNAs, and piRNAs? What are their roles?
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
30 Terms
What is the model organism that played a key role in the discovery of RNAi?
a. Saccharomyces cerevisiae
b. Caenorhabditis elegans
c. Xenopus laevis
d. Drosophila melanogaster
b. Caenorhabditis elegans
Which enzyme cleaves long, double-stranded RNA into short pieces of double-stranded RNA?
a. RISC
b. Drosha
c. Dicer
d. Argonaute
c. Dicer
What do PAZ domains do?
a. Bind to single-stranded 3' ends of RNA
b. Repress translation
c. Cleave RNA
d. Dissociate double-stranded RNA
a. Bind to single-stranded 3' ends of RNA
Which miRNA is critical for timing the L1 to L2 transition in C. elegans?
a. miR-4
b. RNAi-4
c. lin-4
d. dsRNA-4
c. lin-4
What is encoded by the genes that piRNAs target?
a. Recombinases
b. Transposases
c. Polymerases
d. Ligases
b. Transposases
How was RNAi discovered?
Nobel prize in 2006 to Andrew Fire and Craig Mello
Which enzyme cleaves double-stranded RNA introduced from outside the cell?
A. Dicer
B. Drosha
C. RISC
D. Argonaute
A. Dicer
Which complex then binds the dsRNA?
A. Dicer
B. Drosha
C. RISC
D. Argonaute
C. RISC
Which component of the complex holds the siRNA and cleaves the target mRNA?
A. Dicer
B. Drosha
C. RISC
D. Argonaute
D. Argonaute
Which domain of Argonaute binds to the siRNA?
A. SH2 domain
B. Recognition helix
C. PAZ domain
D. RNAse domain
C. PAZ domain
When Argonaute cleaves an mRNA, that mRNA can no longer be translated. This is true even when the RNA is cleaved outside the protein-coding region, e.g. in the 3’ UTR. Why is this the case?
A) Cleavage of the 3’ UTR prevents the mRNA from being transported from nucleus to cytoplasm.
B) Eukaryotic ribosomes translate mRNA from 3’ to 5’, so cleavage in the 3’ UTR prevents the ribosome from reaching the protein-coding sequence.
C) Cleavage of the 3’ UTR disrupts the normal pattern of splicing.
D) Eukaryotic ribosomes only translate mRNAs that have both a 5’ methyl-G cap and a 3’ poly(A) tail.
D) Eukaryotic ribosomes only translate mRNAs that have both a 5’ methyl-G cap and a 3’ poly(A) tail.
Where do miRNAs come from?
A. They are introduced from outside the cell
B. They are in the germline
C. They are integrated into the genome via transposons
D. They are encoded within genes in the genome
D. They are encoded within genes in the genome
What is required for a stretch of RNA sequence to be processed into a miRNA?
A. It is complementary to a target mRNA
B. It forms a hairpin loop with characteristic bulges
C. It is located in an intron
D. It is transcribed by Polymerase II
B. It forms a hairpin loop with characteristic bulges
Typical test question: Both siRNAs and miRNAs can activate RNA interference, and they use many of the same proteins to do so.
Which of the following is not a common feature of these two pathways?
A) PAZ domains
B) RISC
C) Drosha
D) Argonaute
E) Dicer
C) Drosha
Which of the following is not a common feature of the activity of Drosha and Dicer?
A. They cleave both strands of the double-stranded RNA
B. They leave 3’ overhangs at the cleavage site
C. They act in the nucleus
D. They recognize their targets by shape
C. They act in the nucleus
What is one way that miRNAs do NOT reduce gene expression?
A. Degradation of target mRNAs
B. Sequester target mRNAs in the nucleus
C. Inhibit translation of target mRNAs
D. Suppress gene transcription through chromatin modification
B. Sequester target mRNAs in the nucleus
Which miRNA controls the transition from L1 to L2 in C. elegans?
A. lin-4
B. lin-14
C. let-7
D. lin-41
A. lin-4
How does lin-4 control the transition from L1 to L2 in C. elegans?
A. It reduces expression of Lin-41
B. It reduces expression of Lin-14
C. It increases expression of Lin-41
D. It increases expression of Lin-14
B. It reduces expression of Lin-14
How were the existence and roles of lin-4 and let-7 discovered?
A. Through microarrays for genes that change expression level during development
B. Through genetic screens for mutations that affect development
C. Through transgenic experiments testing random RNA sequences
D. Through biochemical experiments searching for things that bind to the 3’UTR of the lin-14 mRNA
B. Through genetic screens for mutations that affect development
What were the inhibitor molecules that stopped P element transposition in P-strain flies?
A. miRNAs
B. siRNAs
C. piRNAs
D. dsRNAs
C. piRNAs
In which cell types do piRNAs function?
A. Germ cells
B. Somatic cells
C. Neurons
D. Blood cells
A. Germ cells
what did Andrew Fire and Craig Mello discover in 1998?
RNA interference (RNAi): that the expression of any specific gene could be strongly and reversibly inhibited by introducing a dsRNA containing a base sequence complementary to that gene’s mRNA
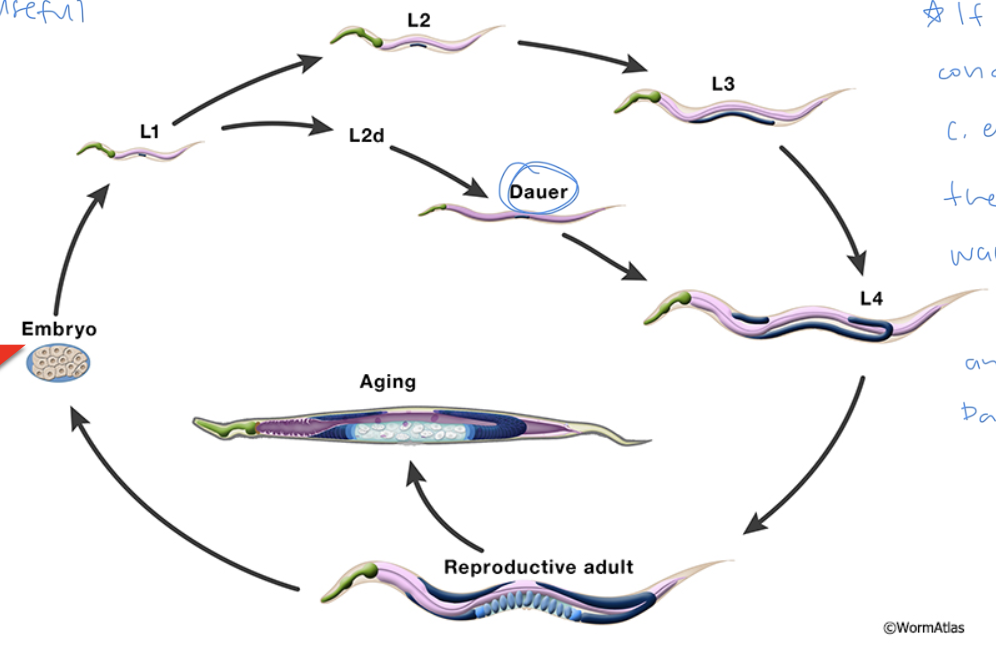
C. elegans life cycle
embryo → L1 → L2 → L3 → 4 → reproductive adult → aging
if environmental conditions are bad during L1, C. elegans can enter the Dauer phase and wait there until conditions get better, and then it can exit Dauer phase and enter L4
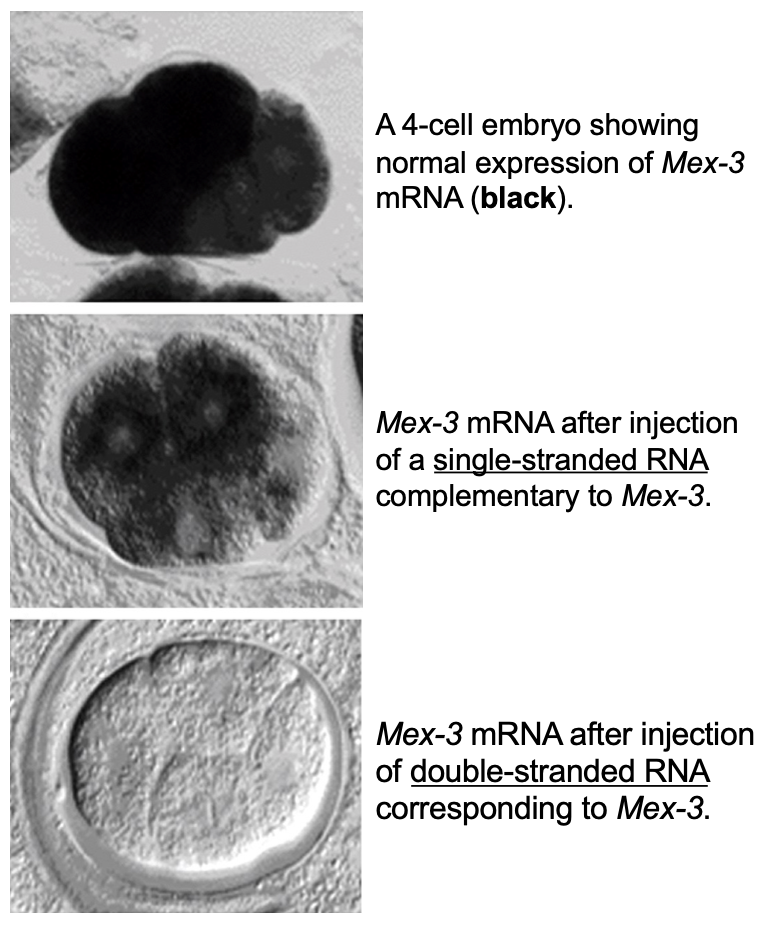
what happens if c. elegans embryos were injected with a ssRNA complementary to a particular mRNA? with a dsRNA?
ssRNA → modest reduction in gene expression
dsRNA → complete eliminated of gene expression
this effect is known as RNA interference (RNAi)
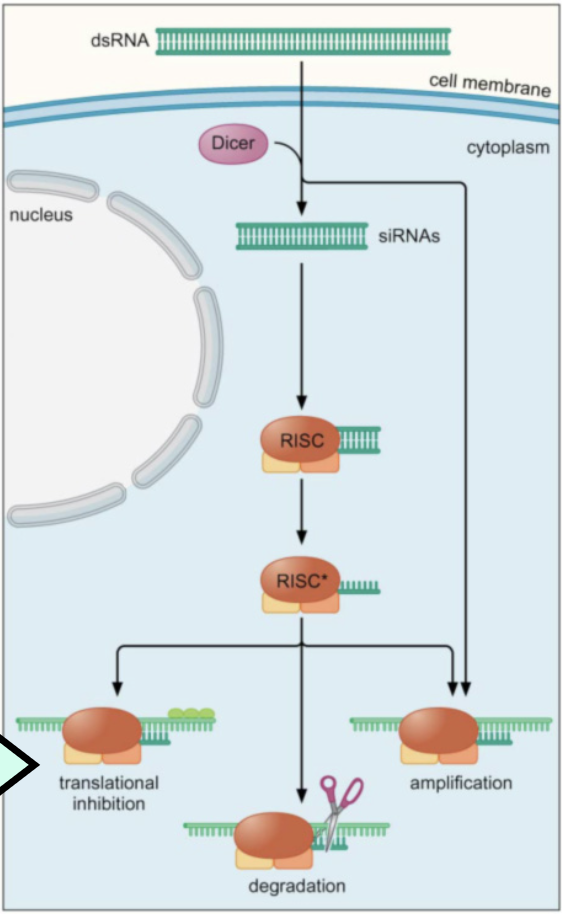
RNAi with dsRNA/siRNA
dsRNA introduced from outside the cell
enzyme Dicer cleaves the dsRNA randomly into 22 bp fragments → siRNAs (small-interfering RNAs)
protein complex RISC binds to ds-siRNA, causing it to denature
siRNA:RISC complex can:
bind to complementary mRNA and prevent translation
OR
cleave and degrade target mRNA
OR
amplify and produce more siRNA molecules
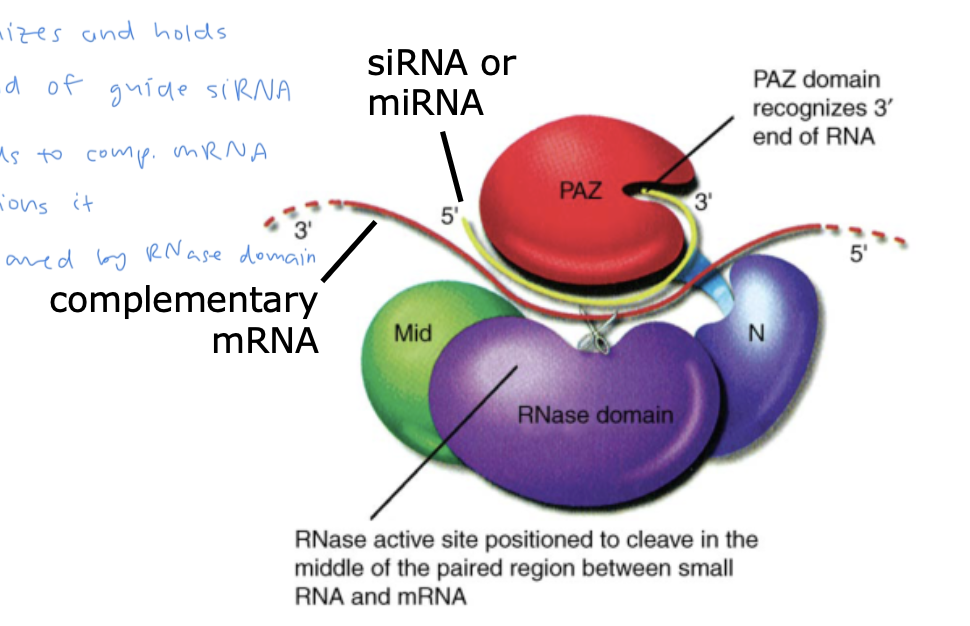
RISC
a large complex containing multiple proteins, including RNase Argonaute
Argonaute has a PAZ domain which:
recognizes and holds onto the 3’ end of the guide siRNA
siRNA binds to complementary mRNA and positions it
mRNA is cleaved by Argonaute’s RNase domain
(picture is of Argonaute)
RNAi with miRNAs
endogenous dsRNA encoded in the organism’s own genome
transcribed by RNA Pol II
primary transcript is both capped and polyadenylated
each functional miRNA arises from a stem loop in the secondary structure of the primary transcript → it is cut out of the primary transcription by two RNases
the first RNase, Drosha, binds to and cleaves the pri-miRNA within the nucleus
Drosha cleaves a single phosphodiester bond on either side of the stem loop, leaving a 3’ overhang and releasing the stem loop, which is now known as a pre-miRNA
cleavage sites are determined by the shape of the stem loop, not the base sequence
the pre-miRNA is transported to the cytoplasm, where its terminal loop is cleaved off by a second RNase, Dicer
Dicer has a PAZ domain (structurally homologous to the PAZ domain found in Argonaute) that recognizes the 3’ overhand of the pre-miRNA
Dicer measures ~22 bases from each Drosha cute site, and cleaves the phosphodiester bonds at those two locations, which also leaves a 3’ overhang
RISC binds to miRNA
miRNA:RISC complex can:
inhibit translation
OR
cause degradation
OR
enter the nucleus and affect gene expression via chromatin remodeling
role of microRNAs (miRNAs) in gene regulation
the human genome encodes >5000 distinct miRNAs
over 60% of human protein-coding genes are regulated by one or more miRNAs
mutations in miRNA genes can result in disease
some act as ‘tumor suppressors’ → their loss results in the over-expression of genes that can cause cancer
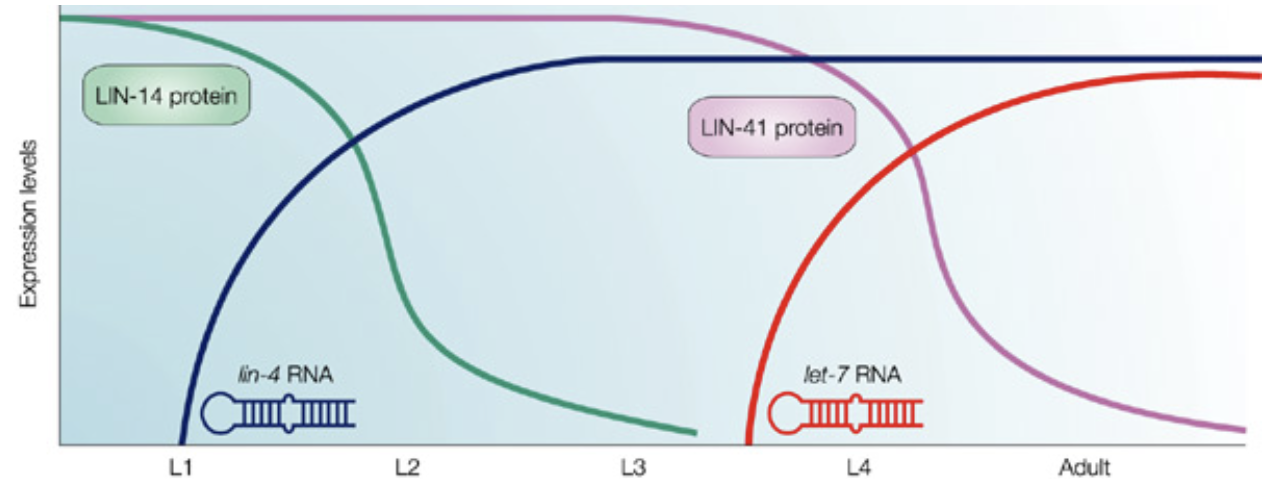
lin-4
in C. elegans, the gene lin-4 produces a 25 base miRNA
this lin-4 miRNA can base pair with any of 7 nucleotide sequences in the 3’ UTR the lin-14 mRNA
this base-pairing does not need to be perfect
when lin-4 miRNA anneals to lin-14 mRNA, expression of lin-14 protein is reduced
lin-14 protein expression is high during embryogenesis → drops after L1 phase due to presence of lin-4 RNA which targets lin-14 protein
similar pattern with lin-41 protein and let-7 RNA in between L3+L4 phase
lin-4 mutants: many adult structures are never made → unable to lay eggs
lin-14 mutants: some adult structures are made too soon → worm is too small and malformed
piRNAs (piwi-interacting RNAs)
a class of short regulatory RNAs
like miRNAs, piRNAs are transcribed from within the organism’s own genome
primarily in germ line cells, where they interact with a germ cell protein called Piwi
within germ line cells, piRNAs and Piwi act together to silence any transposons that share base pair sequence with the piRNA
via DNA methylation and heterochromatin formation around the site of transposon integration