Molecular Bio Unit 1
1/106
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
107 Terms
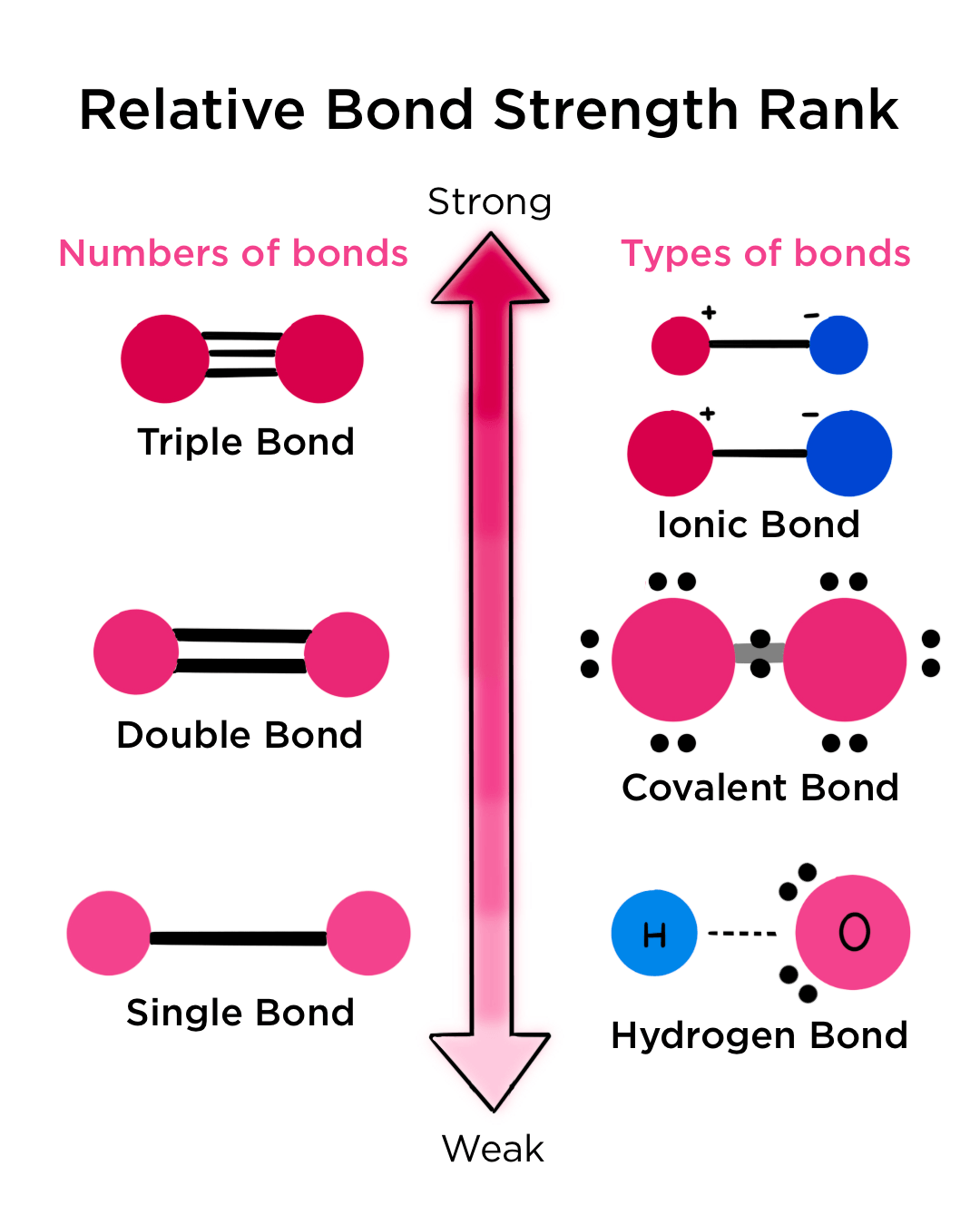
Chemical bonds
two atoms interacting in a way that will make it more likely they will still be interacting in the future
Strong bonds
Stable at physiological temperatures
-covalent bonds
Weak bonds
can be disrupted by certain physiological conditions
Types:
1) Hydrogen bonds
2) Van der Waals
3) Ionic
4) Hydrophobic bonds
Polar molecules
molecules where electrons are not distributed equally (creates a charge difference)
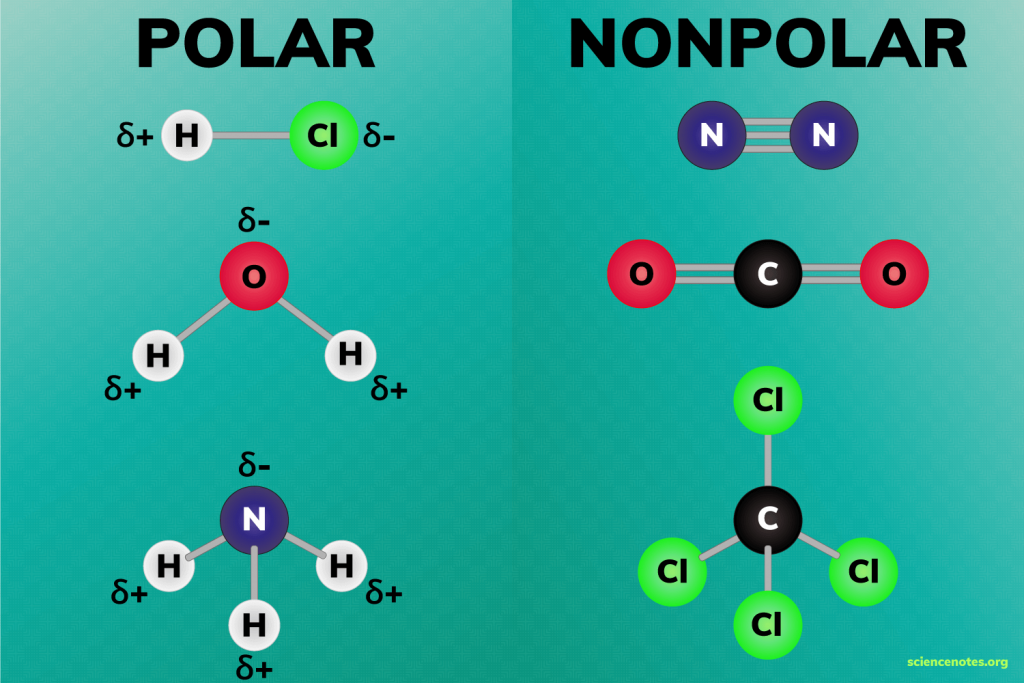
Electronegative
attract electrons
Electropositive
lose electrons
Non-polar molecules
have no dipole moment, balanced
-a molecule where the charges are evenly distributed, meaning there is no significant separation of positive and negative charges. In other words, the molecule doesn't have distinct "poles" (like a positive end and a negative end).
Exp: carbon atoms
Van der Waals forces
nonspecific attraction between 2 atoms in close proximity
Characteristics:
1) Bond strength increases with increasing atom size
2) Repels atoms that are too close
3) Relatively very weak (~1 kcal/mol)
Hydrogen bonds
attraction between an electronegative atom and a electronegative atom
-Not really bonds!
1) Biologically very important
2) Can be formed between any atom that isn’t electrically neutral
3) ~3-2 kcal/mol
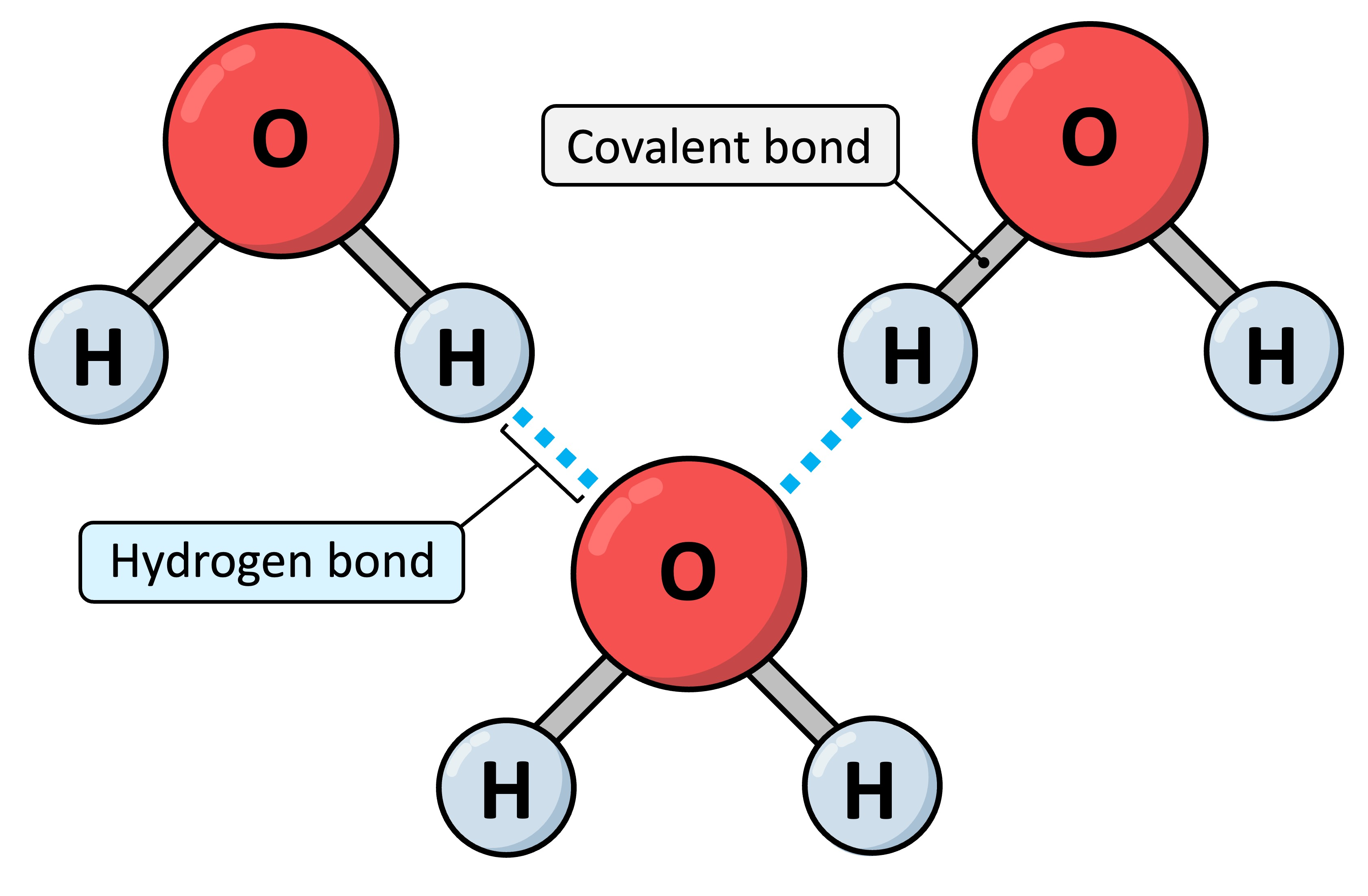
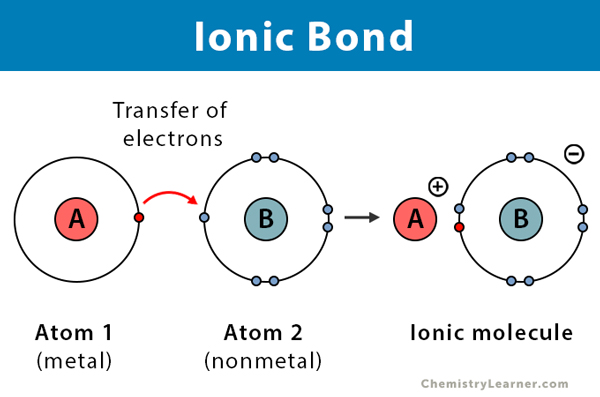
Ionic bonds
charged atoms that are attracted by opposite electrostatic forces
Characteristics:
1) ~5-2 kcal/mol
2) relatively common in molecular bio
3) macromolecules (DNA/RNA/proteins) are usually surrounded by a layer of ions forming ionic & H bonds
Membrane proteins
integral components of cell membranes, playing crucial roles in various cellular processes
-rely on hydrophobic interactions for their structure
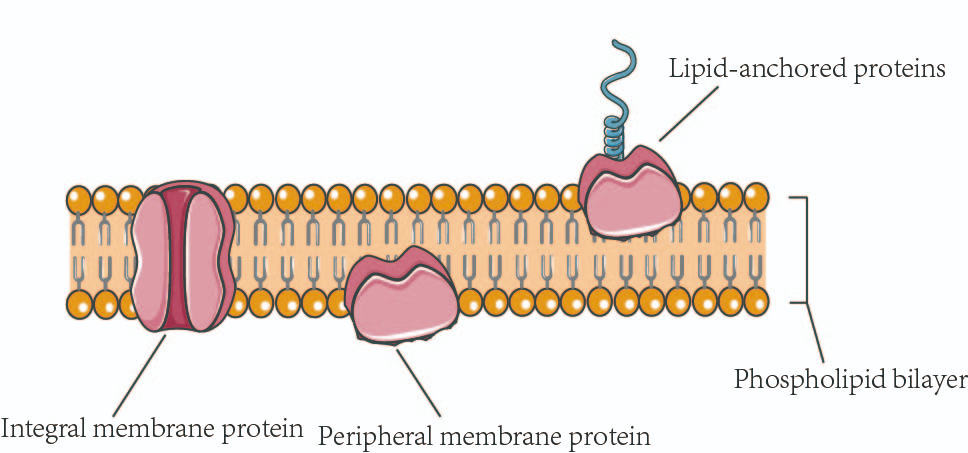
Globular proteins
a type of protein characterized by their spherical or globe-like shape. They are typically soluble in water and play crucial roles in various biological processes.
-require hydrophobic interactions for their structure.
Weak bonds & biological molecules
Guides protein recognition:
-Antibody antigen
-Enzyme-substrate
-DNA-DNA & RNA-RNA
-provides specificity: need many weak bonds to form to get a stable interaction (less chance of error)
very strong at high #s of bonds
Gibbs Free energy
energy required to do work
Activation energy
-energy required to allow a reaction to proceed
-enzymes lower activation energy by stabilizing the transition state and bringing reactants closer together.
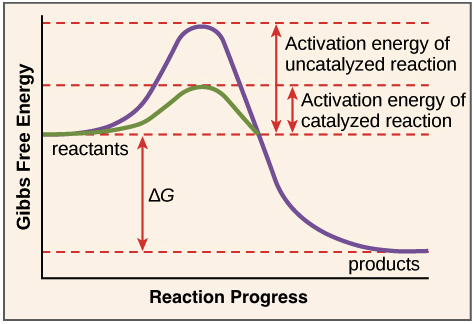
Effect of enzymes
1) Allow exothermic reactions to proceed faster than they “normally” would proceed (in a controlled manner)
2) Allows enzymes to couple endothermic reactions with exothermic reactions to make the whole system energetically favorable
Spontaneous reactions
-exothermic
-will release energy
-will occur on “its own” at some frequency
Non-spontaneous reactions
-endothermic
-requires energy
-would not occur “on its own”
Biological reactions
in a cell, both exothermic and endothermic reactions occur
both kinds catalyzed by enzymes
ATP syhnthesis
formation of high energy bonds that store usable energy
Conservative
a model of DNA replication that produces one new DNA double helix and one old DNA double helix after one round of replication.
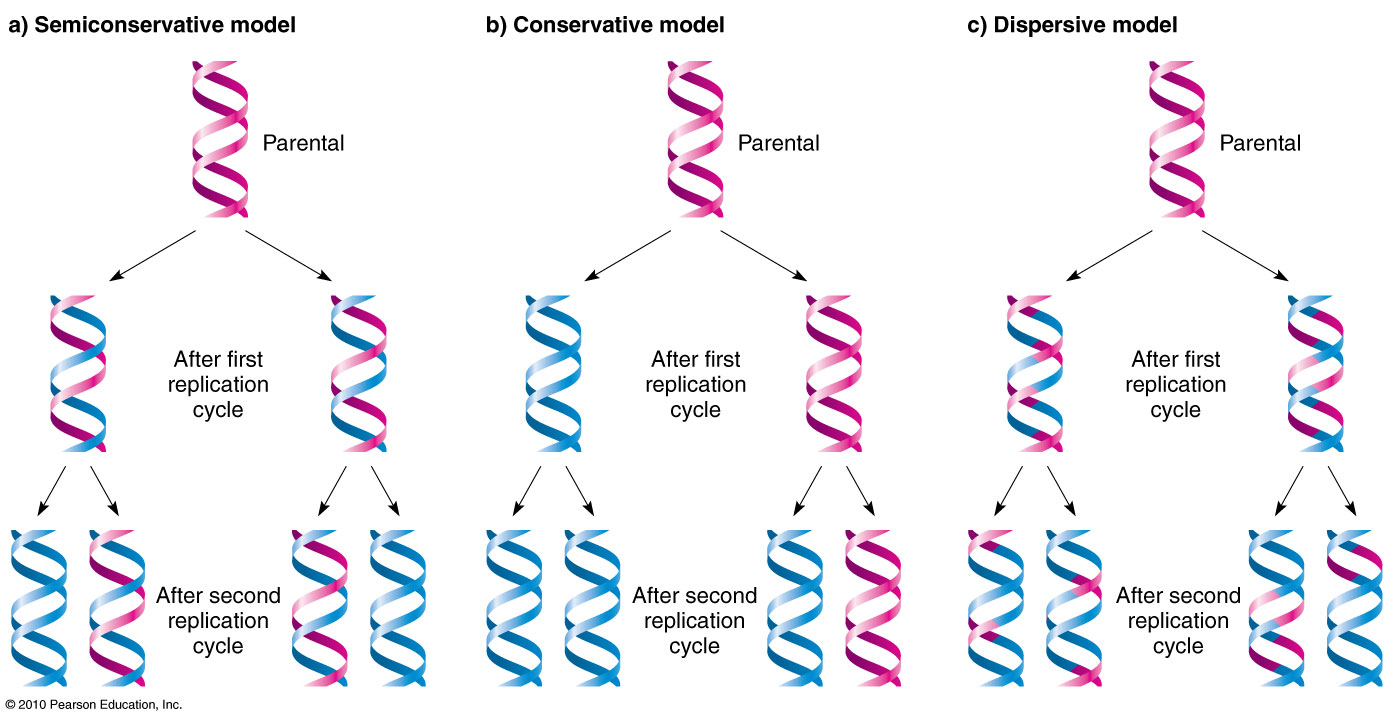
Dispersive
a model of DNA replication that produces DNA molecules that are a mix of new and old DNA.
Semi conservative
a process that copies DNA so that each new DNA molecule has one old strand and one new strand
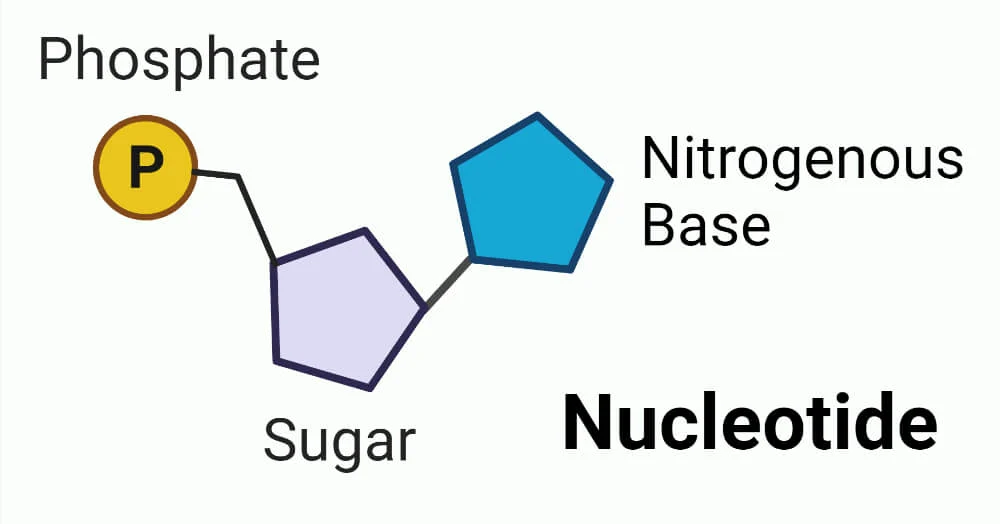
3 components of nucleotides
1) Nitrogenous base
2) Phosphate backbone
3) Sugar (part of backbone)
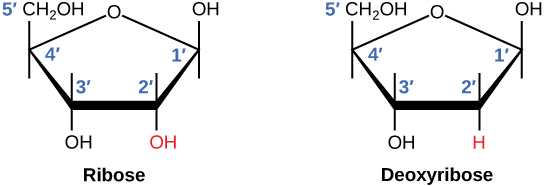
Cyclic 5 carbon sugar
RNA= 2’ carbon has an OH
DNA=2’ carbon has an H
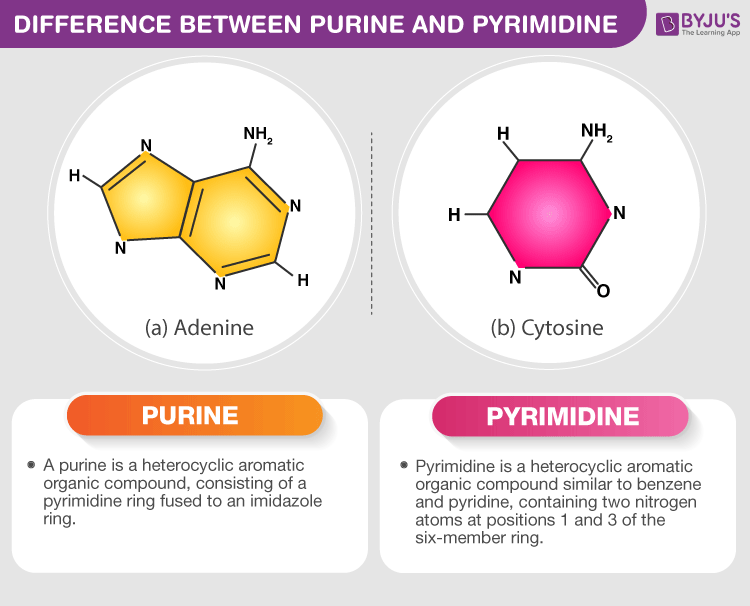
Purines
-adenine & guanine
-two carbon nitrogenous ring bases
Pyrimidines
-Cytosine & thymine
-one carbon nitrogen ring bases
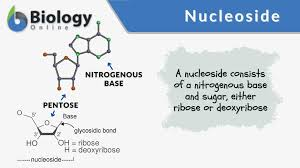
Nucleoside
nitrogenous base + sugar but no phosphate
Pyrophosphate
-2 phosphates still bound together

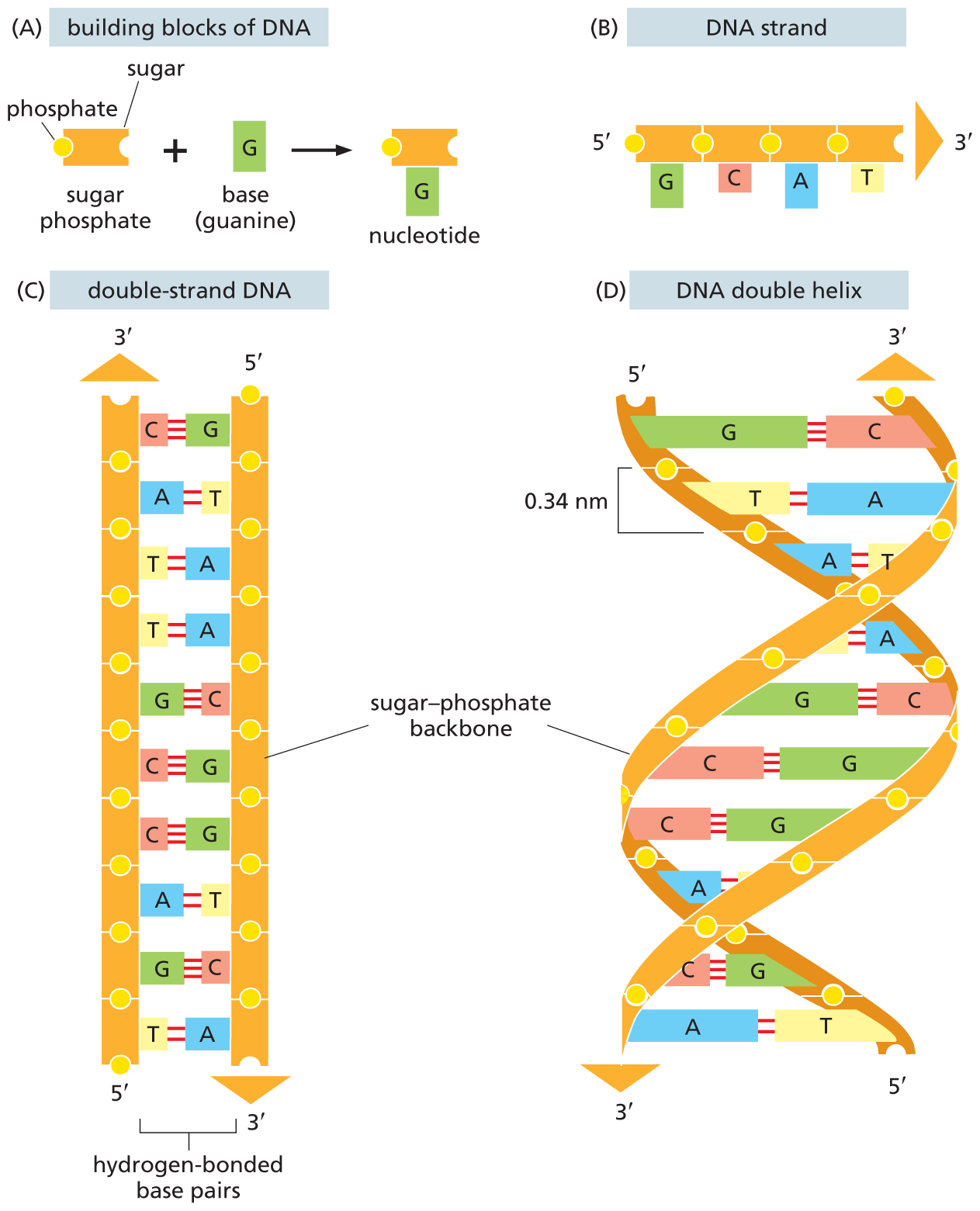
DNA
Deoxyribonucleic acids bound together by phosphodiester bonds
-double stranded
Phosphodiester bonds
occur between the hydroxyl on the 3’ carbon & the phosphate group on the 5’ carbon of the sugar
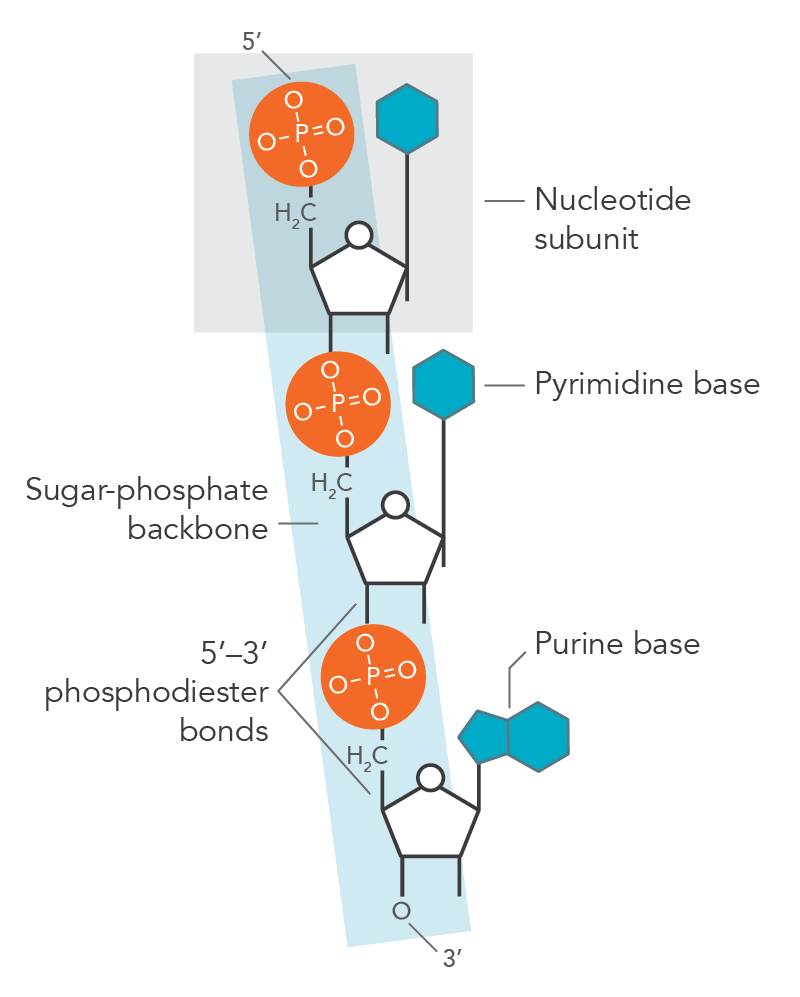
Linkage polarity
5’ —> 3’ polarity
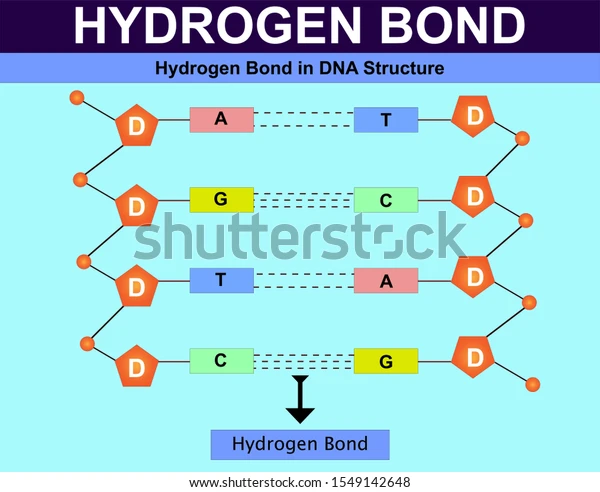
Hydrogen bonds
make up the main force holding two DNA helices together
-each base pair has 2 (A&T) or 3 (G&C) H-bonds between nucleotides
-very weak individually but strong in large numbers
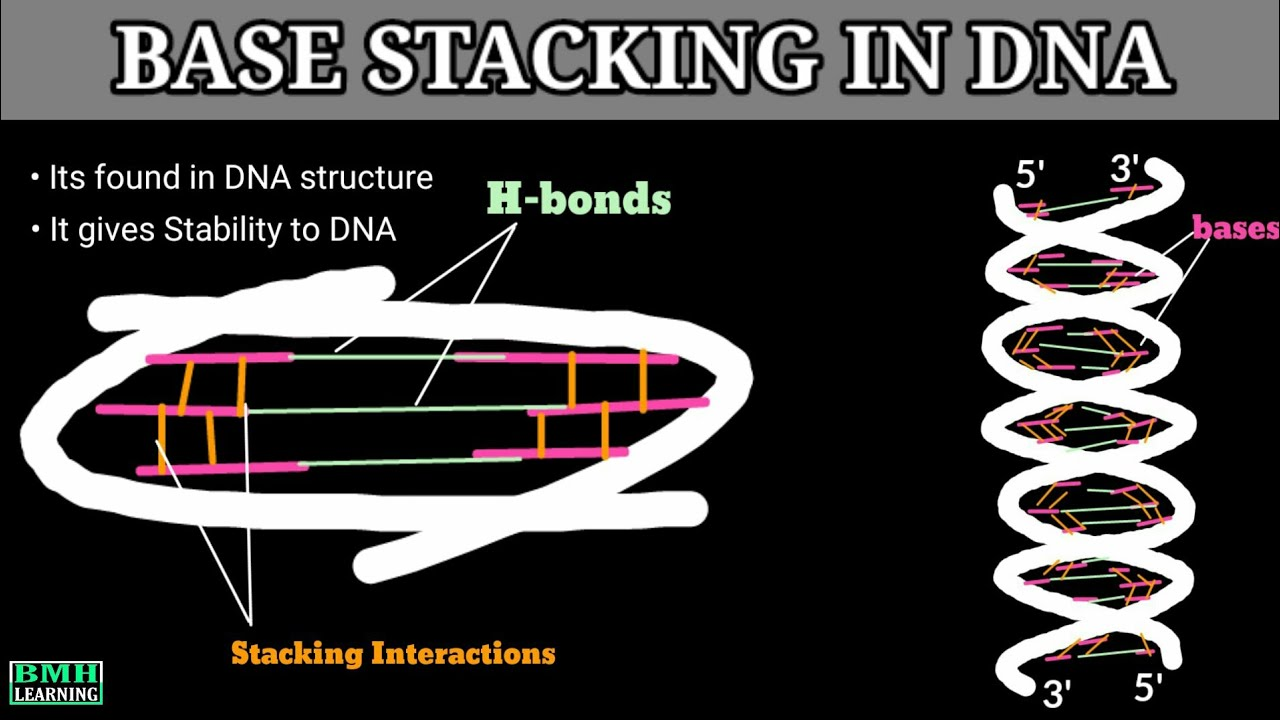
Base stacking interactions
Force that helps stabilize nucleotides stacked in the double helix combination of hydrophobic & Van der Waals forces
Ionic bonds
can form between phosphates & ions in solution
-a type of chemical bond formed when one atom gives away an electron and another takes it. This creates oppositely charged ions (one positive and one negative), and the attraction between these opposite charges holds the atoms together.
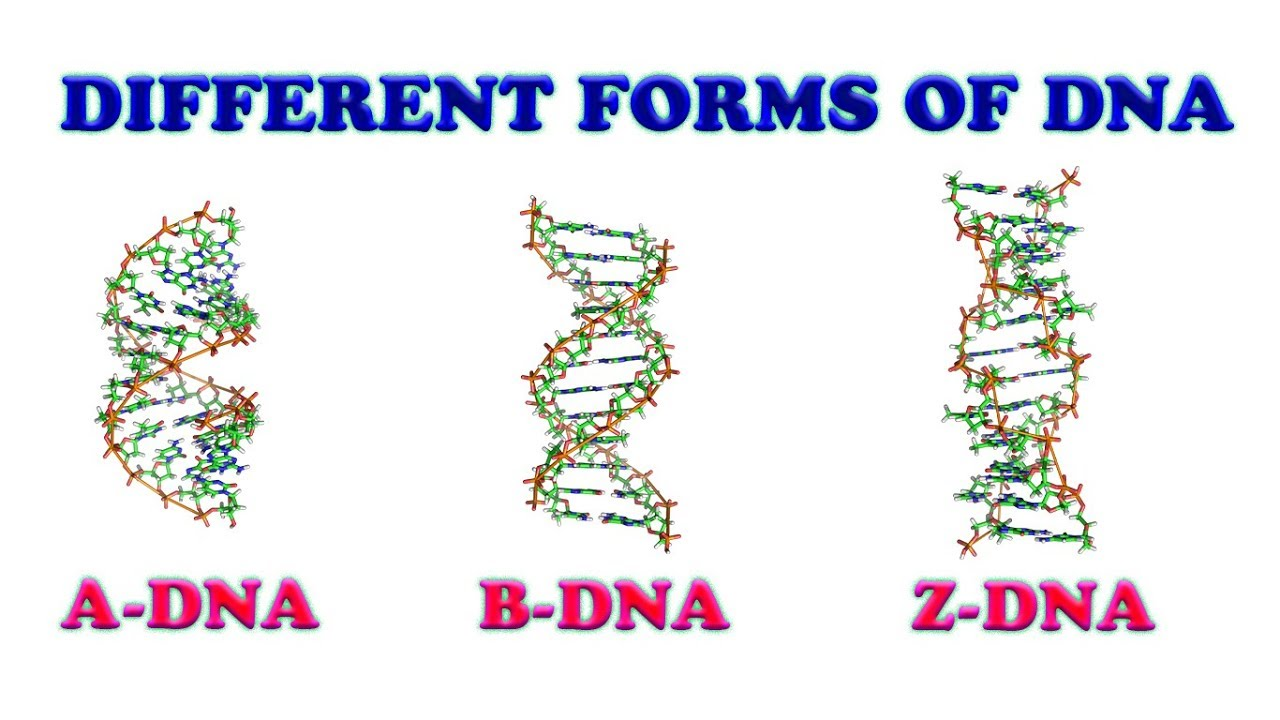
3 forms of DNA
-A, B & Z
B: DNA-DNA dimers, hydrated form of DNA
A: RNA-RNA dimers & RNA-DNA dimers
Z: Thought that the Z form may form spontaneously in G-C rich sequences
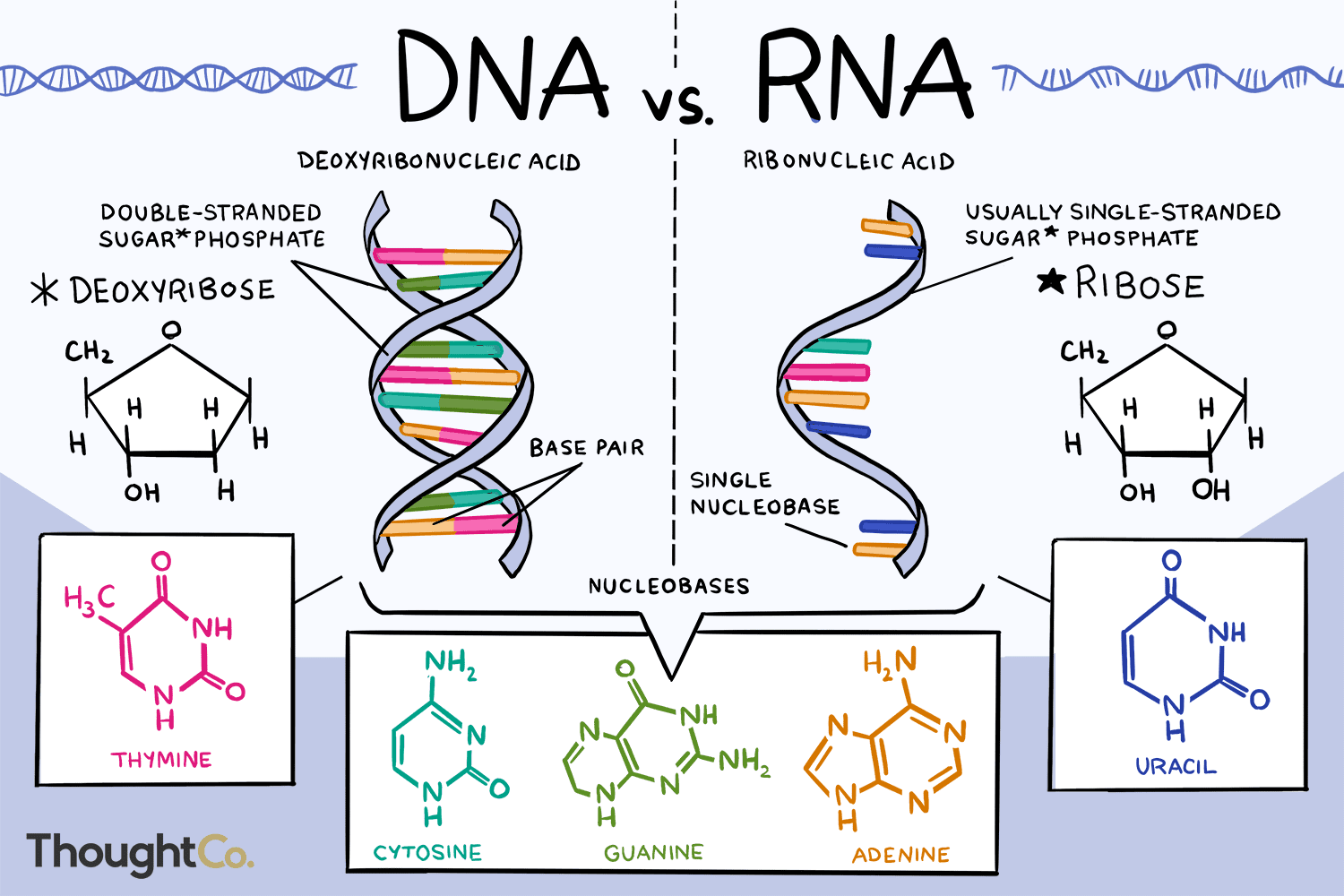
Ribonucleic acid
3 basic types:
-mRNA (messenger)
-tRNA (transfer)
-rRNA (ribosomal)
Differences from DNA:
-uracil instead of thymine
-ribose instead of deoxyribose
-can be found single-stranded
DNA stabilizing bonds
Hydrogen bonds between nucleotide bases (A-T, G-C) stabilize the DNA double helix.
Hydrogen bonds between the phosphate group and water help stabilize the DNA backbone.
Base stacking: Hydrophobic interactions between adjacent nucleotide bases further stabilize the helix structure.
Ionic bonds: Between phosphate groups and ions (like magnesium), which help stabilize the negatively charged DNA backbone.
RNA stabilizing bonds
H-bonds between nucleotides
H-bonds between phosphate & H2O
Base-stacking between nucleotides adjacent
Ionic bonds between phosphates & positive ions
H bonds between 2’ OH & other molecules
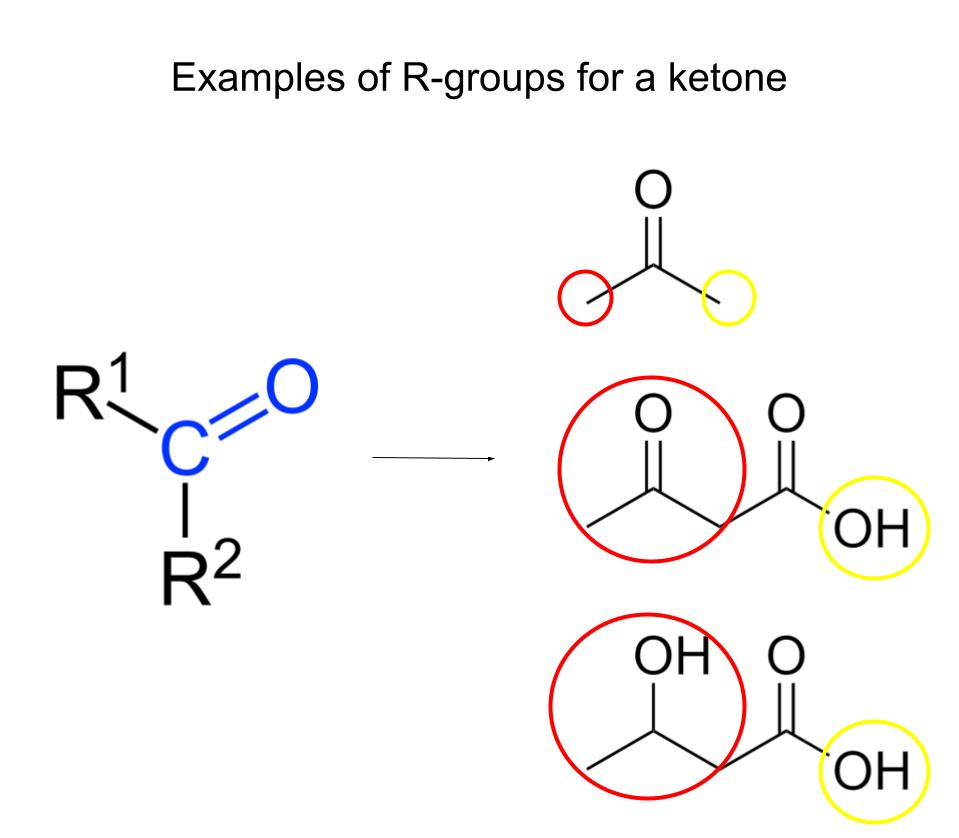
Carboxy & amino groups
-part of the backbone of the protein structure
-can form H-bonds
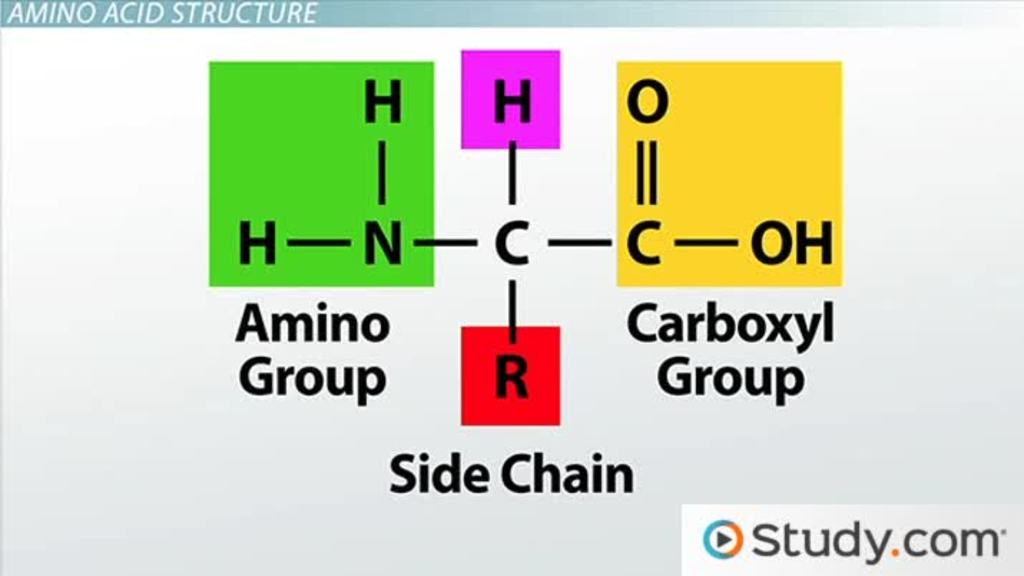
R groups in amino acids
Van der Waals interactions: Large ring-shaped amino acid side chains (R groups) create weak attractions with each other, contributing to protein stability and shape.
Hydrophobic bonds: Hydrophobic amino acids have non-polar R groups that avoid water. These R groups cluster together inside proteins, forming hydrophobic bonds and helping the protein fold.
Hydrogen bonds: Hydrophilic amino acids have polar or charged R groups that attract water. These R groups form hydrogen bonds, stabilizing the protein in aqueous environments.
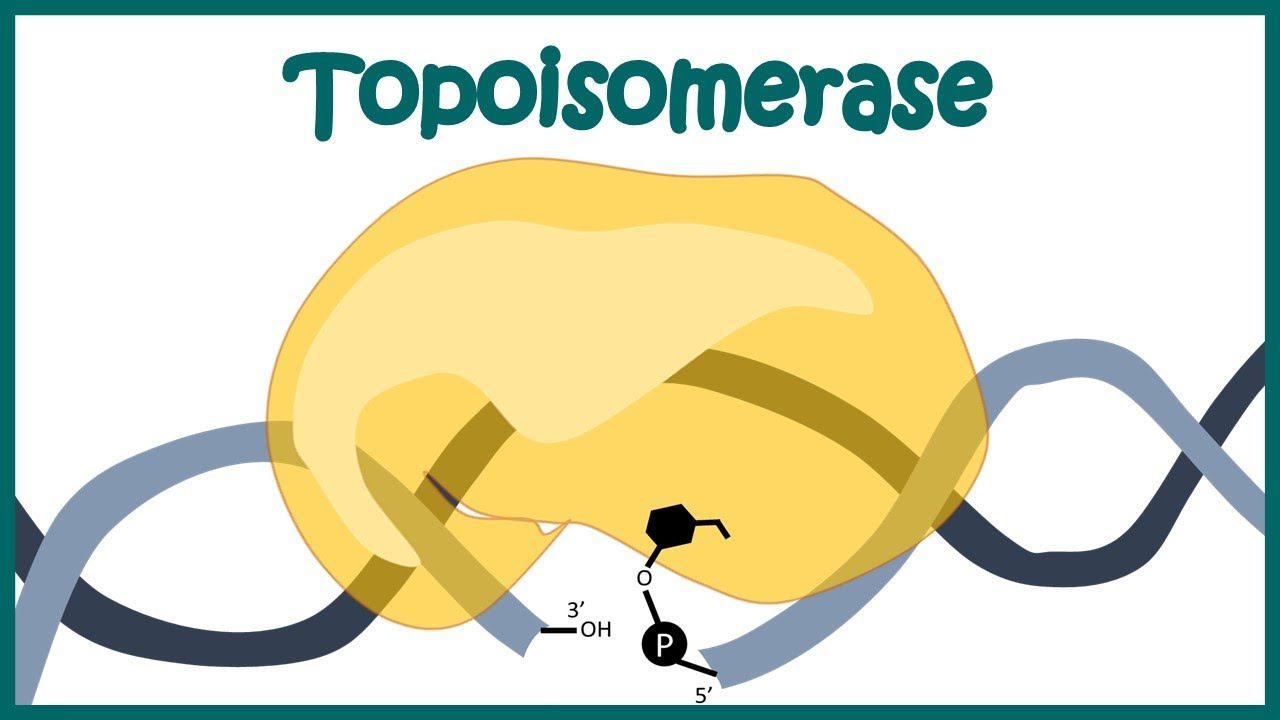
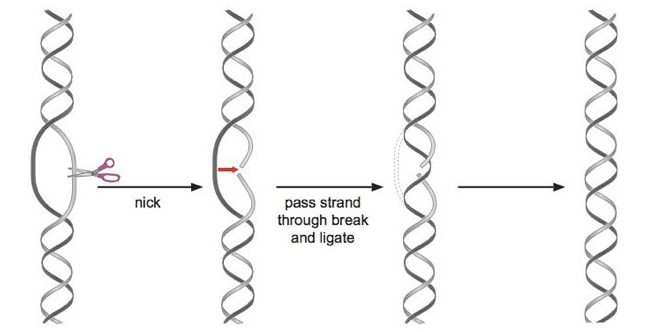
Topoisomerase
Cleaves and reattaches single-stranded DNA.
Does not require ATP or energy input.
Enzymatic activity relies on the close proximity and structural similarity between tyrosine and DNA.
-A specific tyrosine residue in the active site attacks the phosphate backbone of DNA, creating a temporary covalent bond with the DNA. This covalent bond stores energy, allowing topoisomerase to rejoin the DNA without requiring ATP.
No bonds are created or destroyed in the process
Protein primary structure
sequence of amino acids in a polypeptide (protein) chain
20^# of amino acids
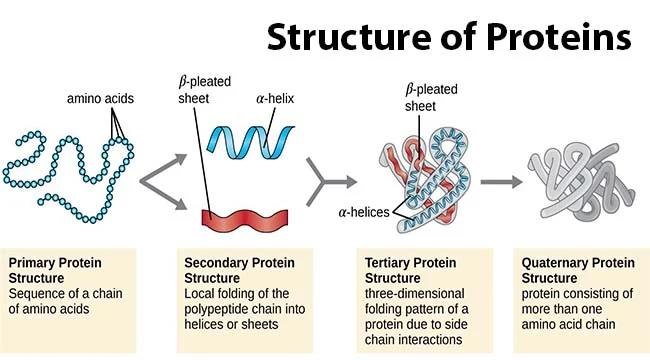
Protein secondary structure
alpha helix & beta sheet (unstructured)
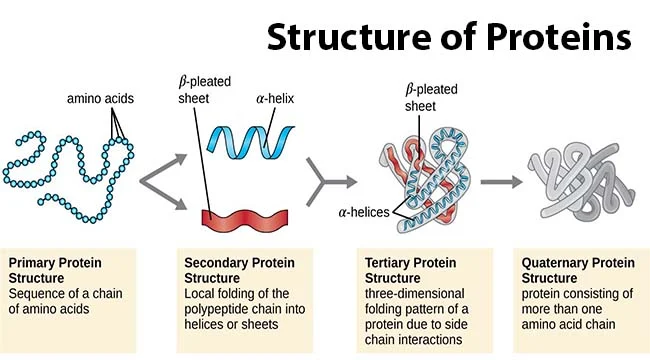
Protein Tertiary structure
folded structures of one polypeptide chain
-infinite
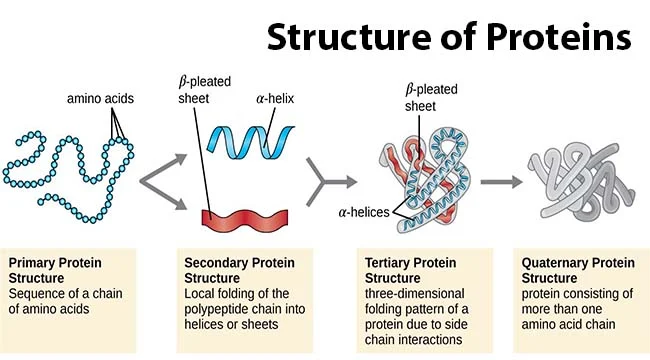
Protein quaternary structure
-protein consisting of more than one amino acid chain
-backbone H-bonds stabilize a-helices & B sheets
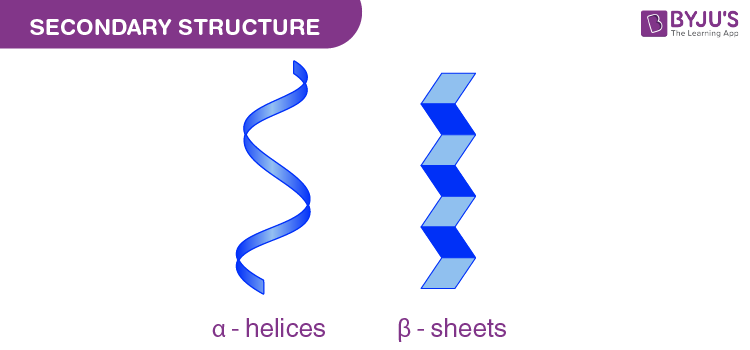
Alpha helices
Stabilization:
Hydrogen bonds between COOH & NH2 in the backbone stabilize the helix.
R-groups positioned outward from the helix.
Right-handed structure.
Helix Destabilizers:
Glycine: Too flexible, allowing free rotation and stressing helix bonds.
Proline: Too rigid, restricting rotation and stressing helix bonds.
Beta sheets
Hydrogen bonds form between COOH (carbonyl) and NH₂ (amide) groups of different β-sheets, stabilizing the structure.
R-groups extend outward, away from the sheet, interacting with the surrounding solution.
Can be parallel (strands run in the same direction) or antiparallel (strands run in opposite directions).
Domain
-a distinct region of a protein that folds together & has a particular function
CAP
Catabolic activating protein
-binds cAMP and then binds DNA
cAMP kinase
binds cAMP & then phosphorylates a substrate
Lac repressor
binds allolactose & then stops binding DNA
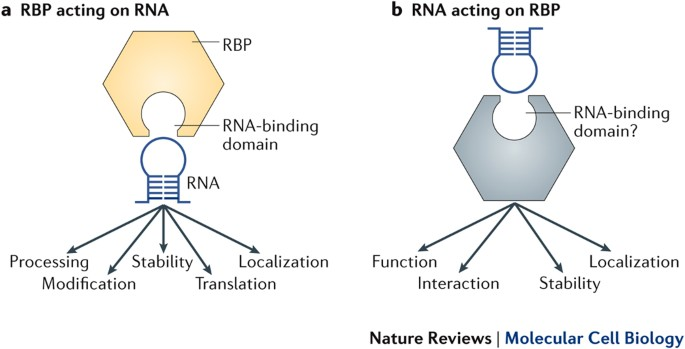
RNA binding domains
Strategy:
RNA-binding proteins contain numerous short RNA binding domains (dozens to hundreds).
Stabilization & Specificity:
Ionic and hydrogen bonds stabilize interactions.
Common Motifs:
Highly diverse!
Many domains with varying patterns within a polypeptide.
Recognition Sequence Impact:
Adapted to bind flexible and dynamic RNA substrates.
Other Domains:
Numerous additional domains contribute to function.
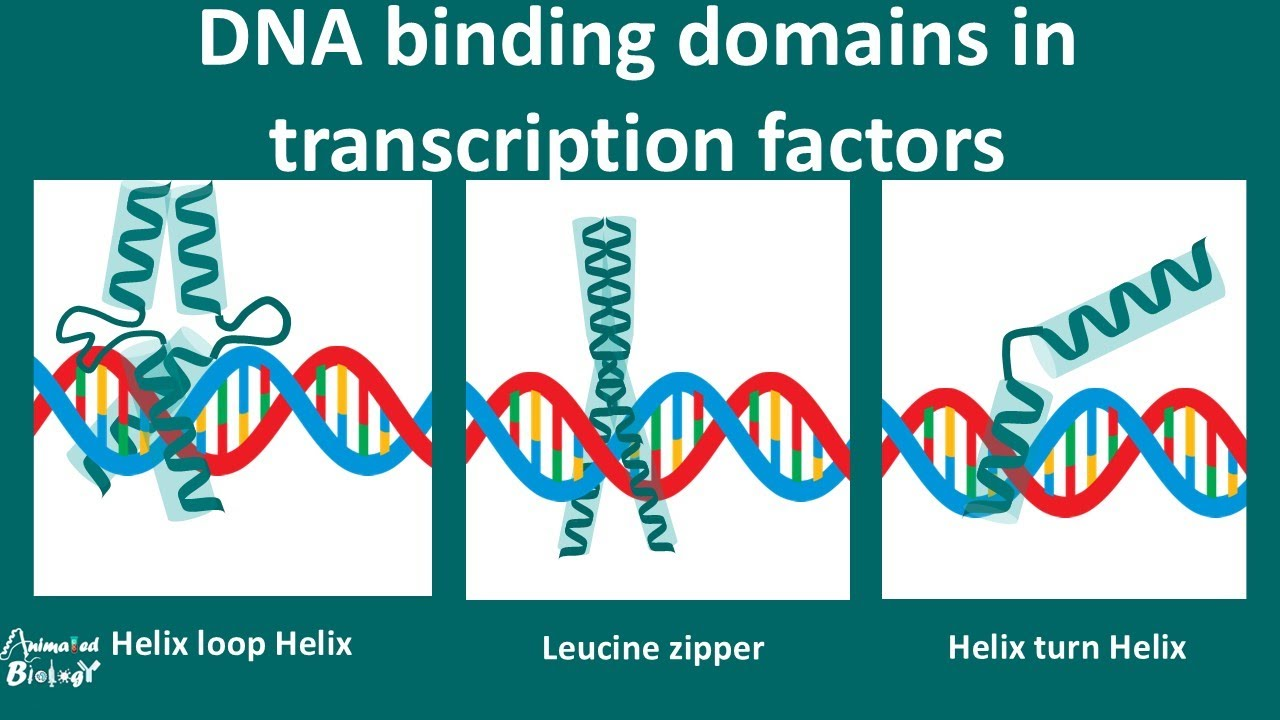
DNA binding domains
Strategy:
Helix-loop-helix DNA binding domain: This refers to a specific structure in proteins that can interact with DNA. It has two helices (spiral shapes) connected by a loop.
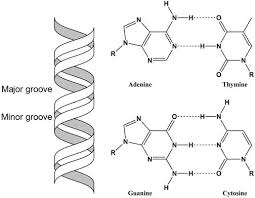
One helix binds the major groove of DNA: The "major groove" is a space in the DNA where certain parts of the DNA structure are exposed, and one helix in the protein forms hydrogen bonds (H-bonds) with it.
The other helix binds the DNA backbone: The second helix in the protein interacts with the backbone of the DNA (the sugar-phosphate chain), using both hydrogen bonds and ionic interactions (electrical attractions between charged groups).
2. Stabilization & Specificity:
Helix #1 (positive amino acids): The first helix contains positively charged amino acids that form hydrogen bonds and ionic interactions with the negatively charged phosphate groups in the DNA backbone.
Helix #2 (polar amino acids): The second helix has polar amino acids that make hydrogen bonds with the nitrogenous bases in the middle of the DNA structure. These bases pair to form the genetic code.
3. Common Motif:
Helix-turn-helix: This is another common pattern found in DNA binding proteins. It involves two helices and a turn (bend) between them, and it is a basic structural feature for proteins that bind to DNA.
4. Impact on Recognition Sequence:
Sequence-specific (“informative”): This means the protein binds to a very specific part of the DNA sequence, like a "lock-and-key" mechanism.
Helix #2 and DNA: The second helix forms hydrogen bonds with the DNA sequence itself.
Helix #1 and nonspecific interactions: The first helix makes weaker, nonspecific interactions with the DNA backbone, meaning it doesn’t recognize a particular sequence but helps stabilize the binding.
5. Other Domains:
Found in many proteins: These DNA binding domains can be found in various proteins, some with only one domain and others with many domains. Each domain helps the protein interact with DNA in different ways.
Prokaryotes
-both circular and linear chromones
-Extrachromosomal elements:
-DNA that exists stably outside of the chromosome (plasmids)
-Ploidy is variable ( E. coli is 1-4N)
Eukaryotes
-linear chromosomes
Ploidy:
1N for haploid
2N for diploid
-can have polypoid cells (>2N)
-Megakaryocytes in humans:
-very large cell found in bone marrow
-produce platelets
-can be up to 16N
Type I Topoisomerase
It cleaves one strand of DNA to relieve supercoiling tension without requiring ATP
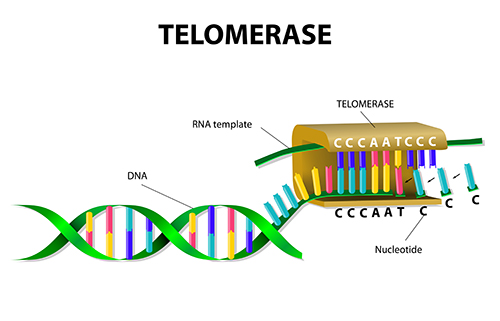
Telomerase
-enzymes that synthesizes telomeres
-a ribonucleoprotein reverse transcription
-an enzyme that has a RNA molecule attached to it
RNA—>DNA
Name the five core histone proteins.
H2A, H2B, H3, H4, and H1 (H1 is the linker histone).
-core has 2 of each
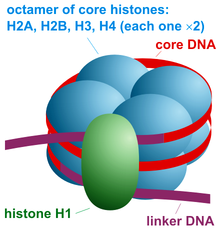
Histone
-positively charged (basic) proteins that interact w/ DNA in a sequence non-specific manner
exps suggest they are not making any/many ionic bonds
-histones are predominantly basic
-BUT doesn’t appear that they make many ionic bonds with DNA
-Actually bind to DNA w/ weaker hydrogen bonds with the minor groove
basic amino acids “balance” the negative charge o the backbone & let the DNA coil more compactly
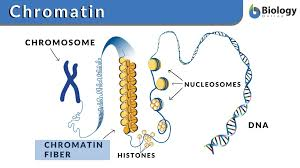
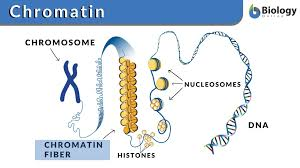
Nucleosome
DNA wrapped around a histone core
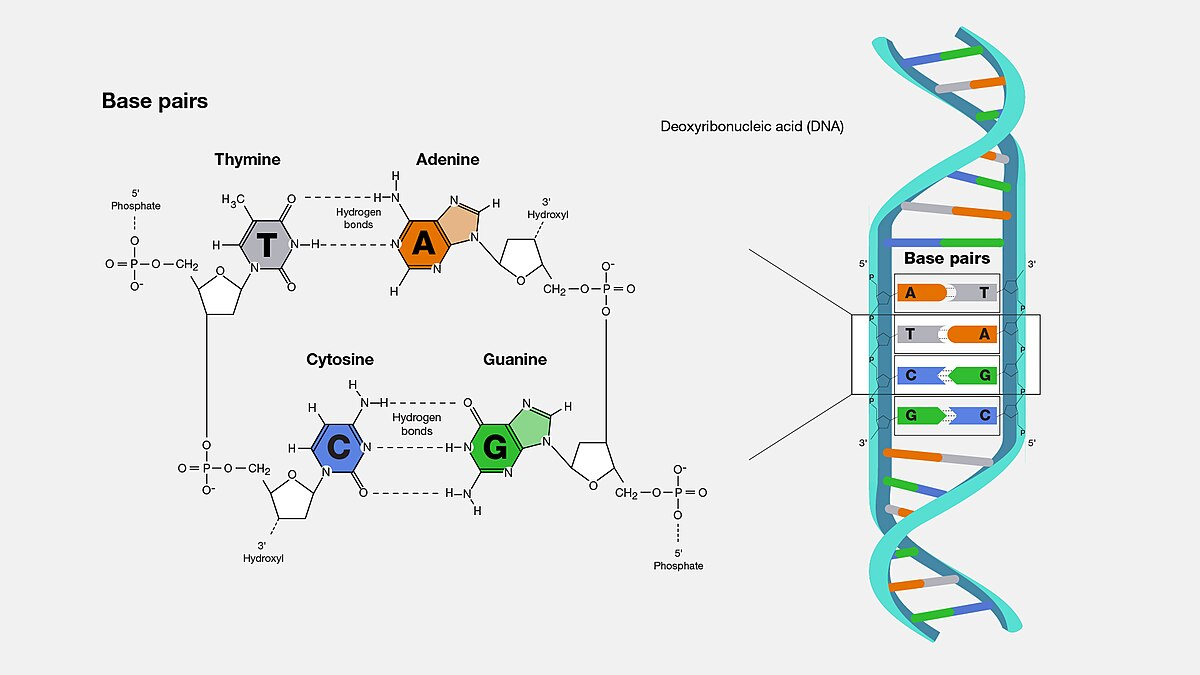
DNA base pairing
Adenine (A) pairs with Thymine (T), and Guanine (G) pairs with Cytosine (C).
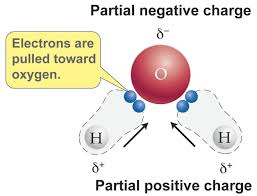
What makes water a polar molecule?
It has partial positive charges on hydrogen and a partial negative charge on oxygen, allowing hydrogen bonding
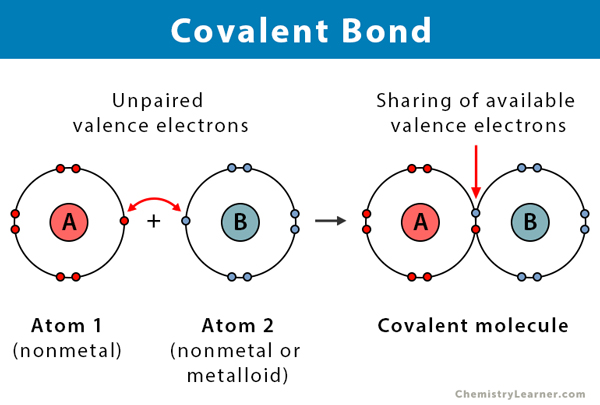
Covalent bonds
-have a larger Gibbs free energy difference, making them stronger and more stable.
How is energy obtained for metabolizing macromolecules?
Through ATP hydrolysis and coupled reactions.
-ATP hydrolysis: exergonic process. It produces ADP (Adenosine diphosphate), Pi (inorganic phosphate) and energy.
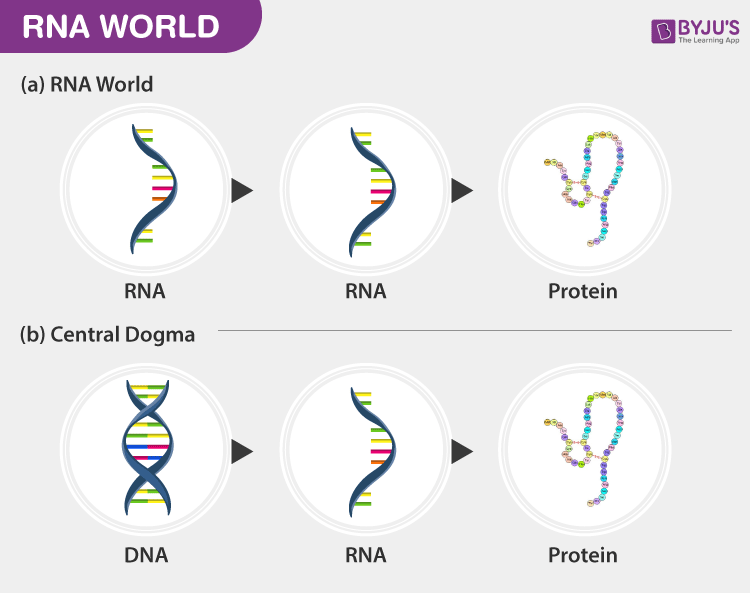
RNA world hypothesis
It suggests that RNA was the first genetic material capable of self-replication and catalysis (the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst)
-in the early stages of life on Earth, RNA molecules were the primary carriers of genetic information and performed catalytic functions, essentially acting as both genes and enzymes, before the evolution of DNA and protein-based life as we know it today; meaning RNA came before DNA and proteins in the evolutionary timeline.
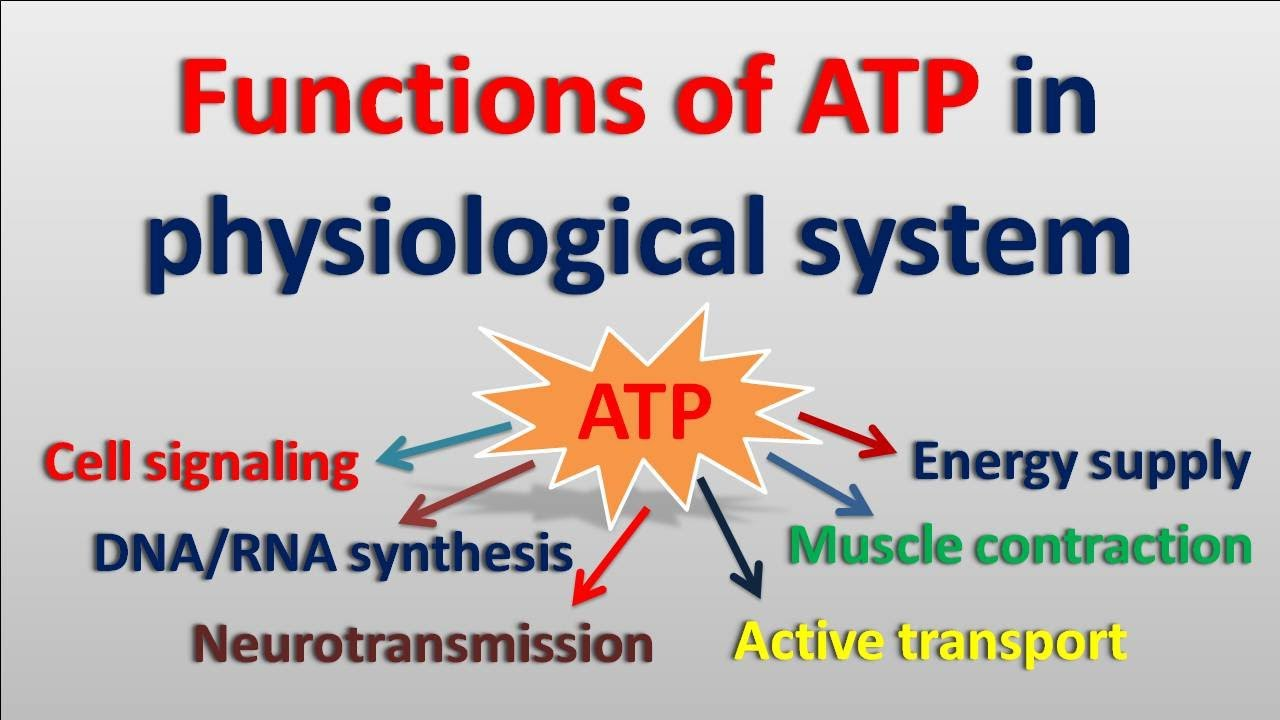
ATP
-serves as the primary energy currency, drives enzymatic reactions, powers molecular motors, regulates signaling pathways, and is used in biosynthetic processes like DNA and protein synthesis.
Major groove of DNA
-Information-rich
-It exposes specific hydrogen bond donors and acceptors for protein binding.
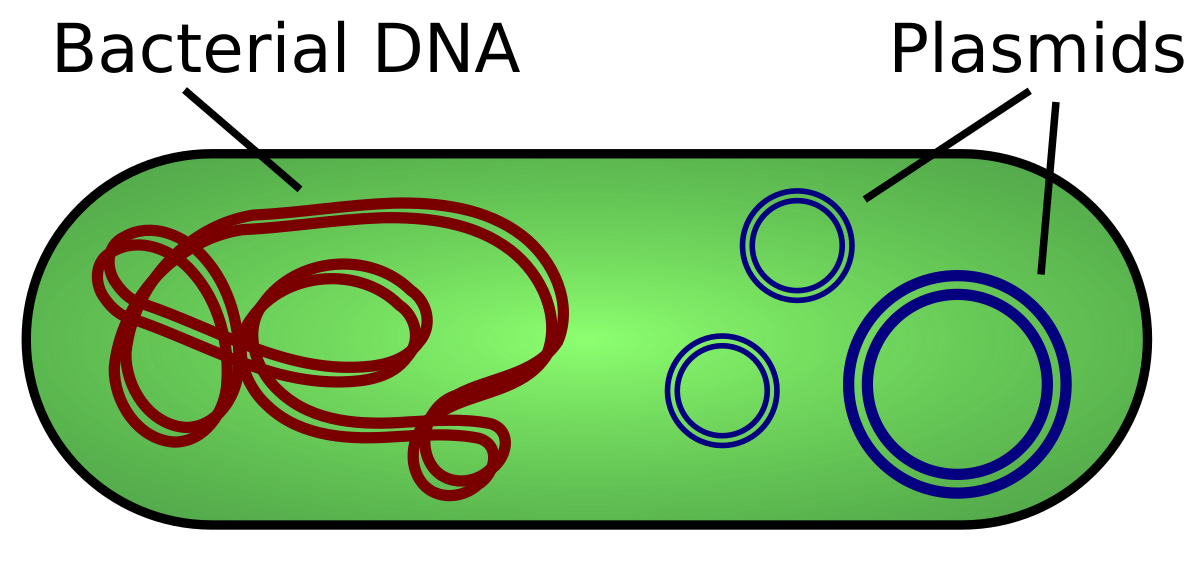
Plasmid
-circular pieces of DNA that replicate & persist in the bacteria cell
Pros:
-provide enzymes for breaking down unusual nutrients.
-provide antibiotic resistance by carrying special genes
Cons:
-require enzymes, nucleotides, and ATP to replicate, which diverts resources from essential cellular functions.
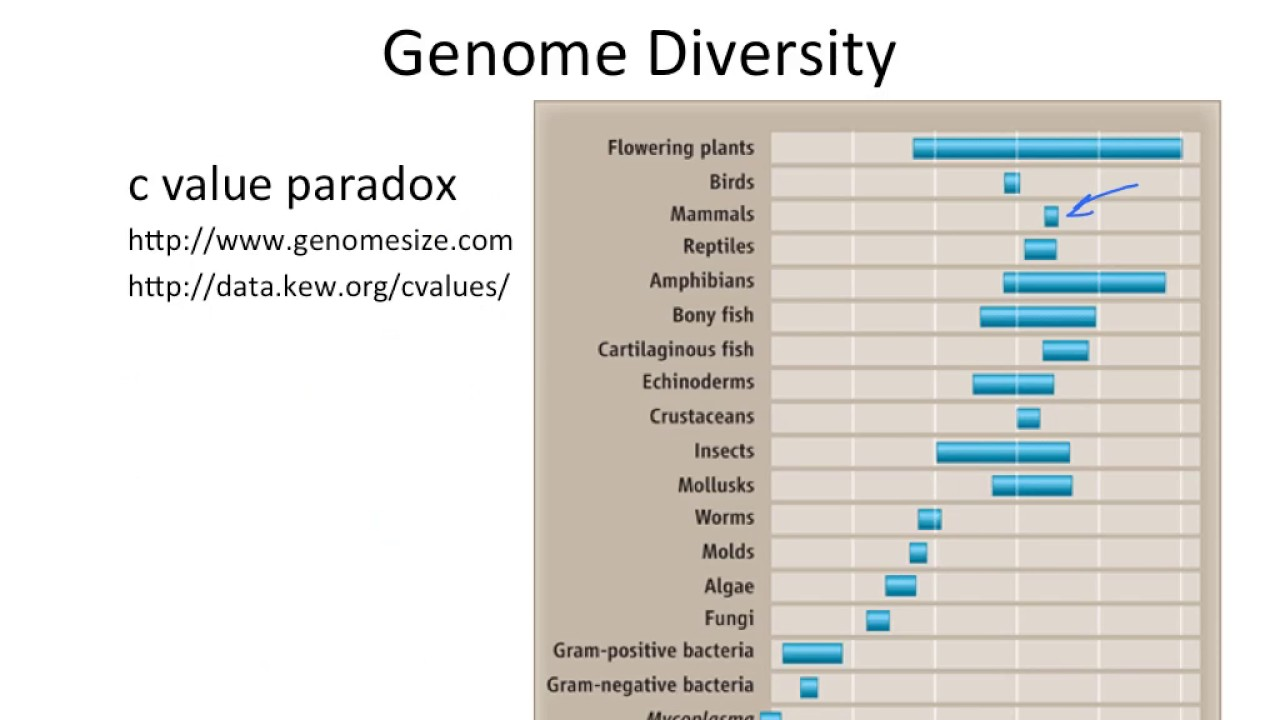
C-value trend in eukaryotes vs. prokaryotes
Only clear trend was prokaryotes have a lot less DNA
C-value= total amount (in grams) of DNA contained in a haploid genome (1 pg= 978 Mb)
C-value paradox
-Genome size does not correlate with organism complexity.
Human genome
-2-5% of the human genome encodes proteins or function
-95%: telomeres, microsatellites (non-genome-wide repeats), functional sequences that aren’t genes, centromeres, regulation regions, genome-wide repeats (transposon)
Alu element
-retrotransposon that are ~300 nt
-most abundant transposon in the human genome
-have more than 1 million copes —> equivalent to 10% of human genome
-thought to increase copies at a very low rate
-no known way to eliminate them
Oxytricha
-protozoa oxytricha genome is full of transposons
-germline: chromosome (has transposons)
-Germline= Changes in the DNA that occur in the reproductive cells (sperm or egg) and can be passed on to offspring
-somatic chromosome (has no transposons)
Somatic=Changes in the DNA that occur in any other cells in the body (non-reproductive cells) and are not passed on to offspring
-during development, transposon genes highly expressed —> flood cell with transposase, which shreds the genome
genome gets reformed as somatic chromosome
experiment suggests transposons now essential for reproduction
Microsatellite DNA
-typically short (2-13 nt) repeats
-exp: 7 x AG repeats @ one locus (one location)
-caused by DNA polymerase “skippping” during replication
-skips ahead —> makes new strand shorter
-skips back—> makes new strand longer
Repetitive DNA w/ known functions:
-telomeres
-centromeres
Prokaryotic chromosome replication
Nucleoid:
-area of a prokaryotic cell where “most” of the DNA is tightly compacted
Supercoiling:
-is how prokaryotes compact their DNA
-is the “super” coiling of DNA into multiple levels of “coiled coils”
Ploidy: is variable depending on location of DNA
-Transcriptionally active & reactive regions of the genome
-loops move out (increase transcription)
-loops move in (decrease transcription)
-more time to replicate the genome than it takes to divide the cell
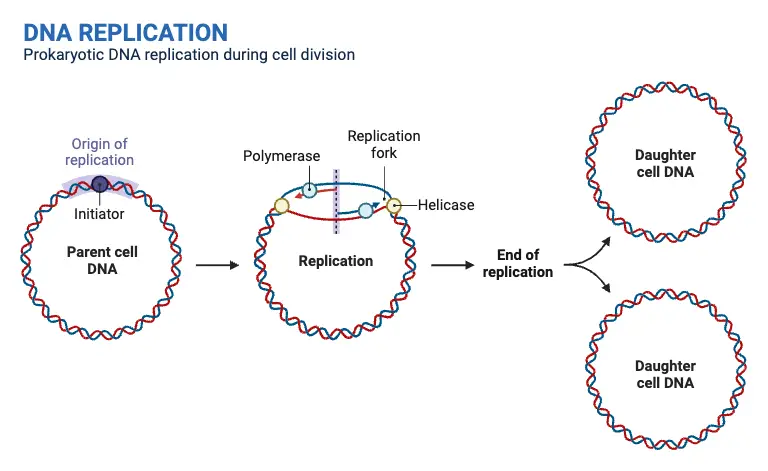
Eukaryotic chromosome replication
Regions of key importance:
Telomere – Protective end caps of chromosomes made of repetitive sequences. They prevent chromosome degradation and fusion but shorten with each replication, contributing to aging.
Centromere – The central region of a chromosome where sister chromatids attach. It is essential for proper chromosome segregation during mitosis and meiosis by serving as the attachment site for spindle fibers.
Origins of Replication (ORI) – Specific DNA sequences where replication begins. Eukaryotic chromosomes have multiple ORIs to ensure efficient and timely DNA replication. Each ORI initiates replication by forming two replication forks that move in opposite directions
Eukaryotic DNA
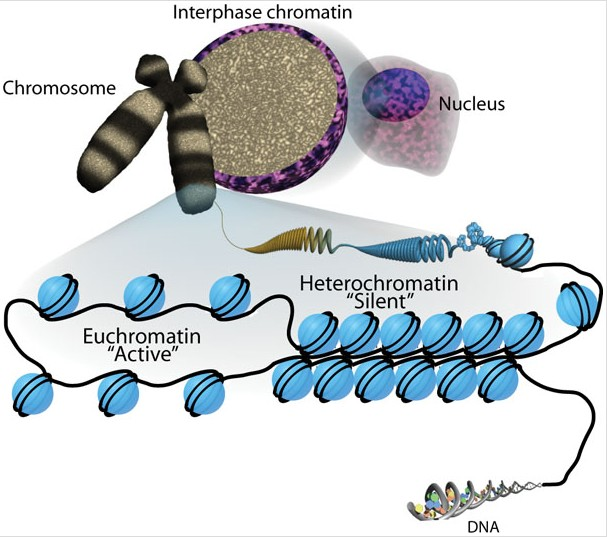
Eukaryotic DNA is very long, so it wraps around histone proteins like thread around a spool.
This wrapping forms structures called nucleosomes, which look like "beads on a string."
Nucleosomes then coil and fold further to form chromatin, which eventually makes up chromosomes when the cell is dividing.
Histone code
-code is either not there for “all organisms” or is too complex
-ones that seem consistent:
-H3 tail: leads to silencing (i.e heterochromatin)
unmodified tail
-H3 tail modified: leads to transcription (heterochromatin + euchromatin) (i.e euchromatin)
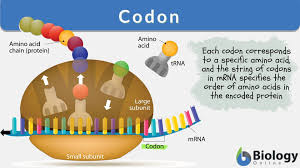
Codons
Sequences of three nucleotides in mRNA that code for a specific amino acid.
Example: AUG (Start codon), UAA (Stop codon).
Phenotype
The physical traits of an organism resulting from genotype and environment.
Example: Eye color, height.
Genotype
The genetic makeup of an organism.
Example: BB, Bb, or bb for eye color.
Haploid
A cell with one set of chromosomes (n).
Example: Gametes (sperm and egg).
Diploid
A cell with two sets of chromosomes (2n).
Example: Most body cells.
Polyploid
Organisms with more than two sets of chromosomes.
Example: Some plants like wheat (hexaploid, 6n).
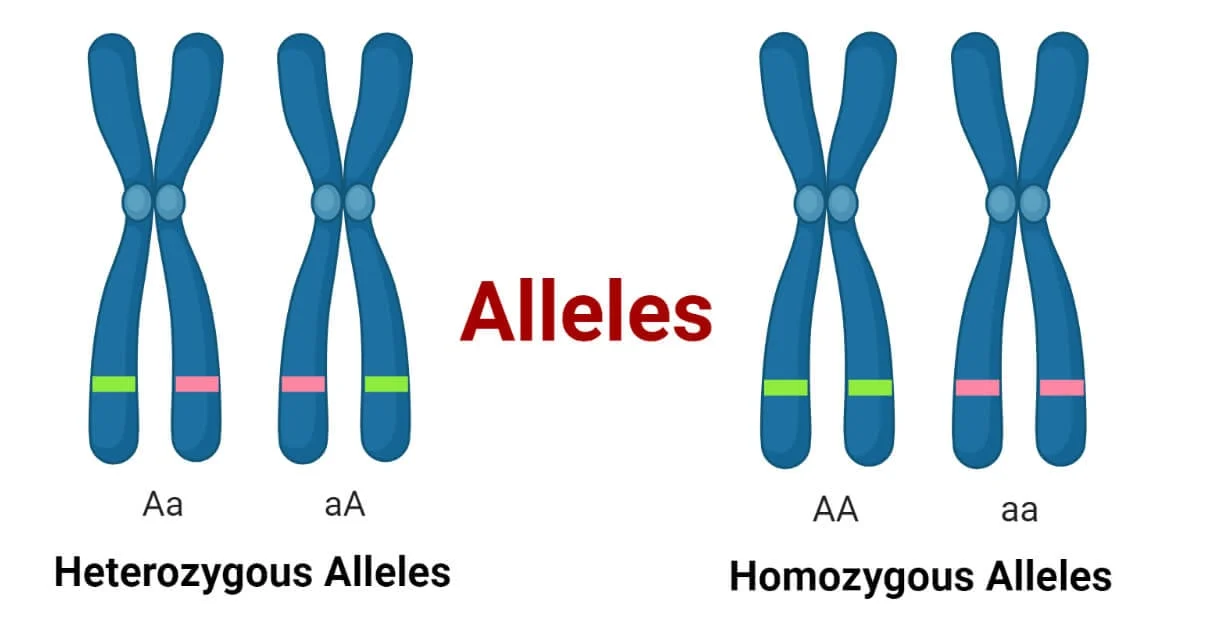
Alleles
Different versions of a gene.
Example: A (dominant) or a (recessive) for eye color.
Wild-type
The most common phenotype in a natural population.
Example: Brown eyes in humans.
Requirements for DNA synthesis
Nucleotides (dNTPs) → The building blocks of DNA that are incorporated into the growing strand.
Ligase & Topoisomerases →
Ligase seals nicks in the sugar-phosphate backbone, joining Okazaki fragments on the lagging strand.
Topoisomerases prevent DNA supercoiling and relieve tension during replication.
Primer → A short RNA or DNA sequence with a free 3’-OH group, required for DNA polymerase to initiate synthesis.
Polymerase → The enzyme that adds nucleotides to the growing DNA strand in a 5’ → 3’ direction.
Template Strand → The single-stranded DNA that serves as a guide for complementary base pairing.
Divalent Cations (Mg²⁺/Zn²⁺) → Essential cofactors that stabilize the DNA polymerase active site and help catalyze nucleotide addition.
DNA polymerase-the palm
Site of catalysis
-active site where new nucleotides are added
-site of primary elements of catalytic site
-binding site for 2 divalent metal ions (Zn²+ or Mg²+)
First metal ion → Lowers the affinity of the 3’-OH group for its hydrogen, making it more reactive for nucleotide addition.
Second metal ion → Stabilizes the negative charge of the pyrophosphate (byproduct of nucleotide incorporation), ensuring efficient catalysis.
Site of 3’ exonuclease proofreading
If a wrong nucleotide is added, the polymerase slows or halts DNA synthesis.
The exonuclease site removes mismatched nucleotides, ensuring high fidelity.
Once corrected, DNA synthesis resumes, reducing mutation rates.
Error rate
-polymerase has about a ~2 in 10^5 error rate alone
-w/ exonuclease domain, error rate decreases to ~1 in 10^7 nucleotides
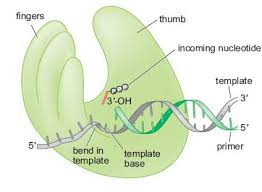
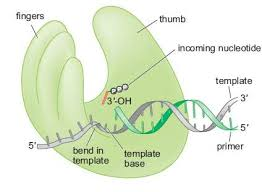
DNA-polymerase-the fingers
-clamping down on a good fit
-responsible for binding nucleotides and positioning the template DNA in the active site.
How does the fingers subdomain work?
The fingers subdomain interacts with the template base and the nucleoside triphosphate (dNTP).
The fingers subdomain positions the template DNA in the active site.
The fingers subdomain is involved in the interactions between the existing template base and the nucleoside triphosphate being inserted
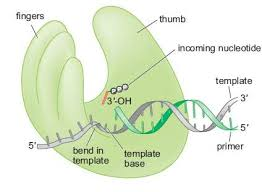
DNA polymerase-the thumb
1. Holding the Synthesized DNA
The thumb domain grips the newly synthesized double helix, ensuring stability.
It maintains electrostatic interactions to help position the DNA correctly for continued synthesis.
2. Interaction with Primer & Template Strand
The thumb keeps the primer and template strands aligned, allowing efficient nucleotide addition.
3. Promoting Processivity
Processivity = The number of nucleotides added before DNA polymerase dissociates.
Higher processivity → faster DNA synthesis
Low processivity → Polymerase dissociates frequently (1 nt per binding).
High processivity → Polymerase stays attached longer (up to 50,000 nts per binding).
Packaging eukaryotic DNA
-the biological problem:
there is too much DNA to fit in the cell!
-the solution:
to organize & package the DNA into tight bundles surrounding a protein core
Nucleosome spacing
Nucleosomes are the basic units of chromatin, consisting of ~200 nucleotides (nt) of DNA wrapped around a histone octamer (H2A, H2B, H3, H4).
Spacing between nucleosomes is about 50 nt, creating a beads-on-a-string structure that helps regulate DNA accessibility.
Each nucleosome core holds ~150–200 nt of DNA, ensuring efficient DNA packaging while allowing regulatory proteins access when needed.
Histone modifications
-histone tails project out of the nucleosome core
-can be modified by other proteins
-can affect nucleoside binding to DNA and chromatin formation
First order of condensation
nucleosomes formed on DNA but no further organization
-”beads on a string” or “10-nm fiber”
-primary state of “euchromatin”
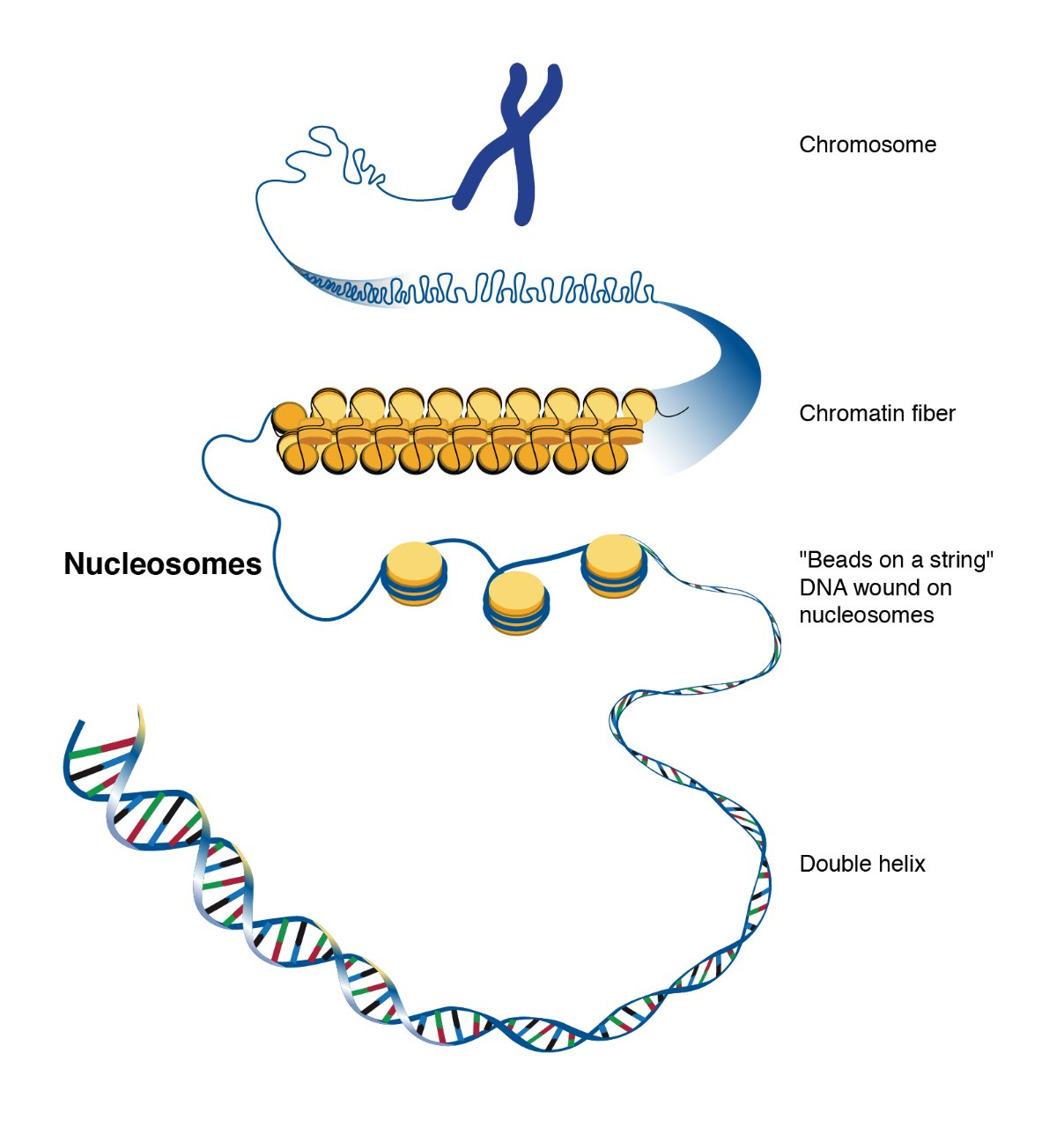
Second order of condensation
-nucleosomes wrapped in a 2nd coil
-called the “30 nm fiber”
-primary state of “heterochromatin”
Higher orders of condensation
-larger super structures found primarily during cell division
-form the “visible” chromosomes
Heterochromatin
more tightly, packed nucleosomes corresponds with transcriptional silencing
-H3 unmodified (meaning it lacks key post-translational modifications like acetylation or methylation)
Acetylation of histone H3 is linked to euchromatin (active, open chromatin). When H3 is unacetylated, chromatin remains tightly packed and transcriptionally silent, characteristic of heterochromatin.
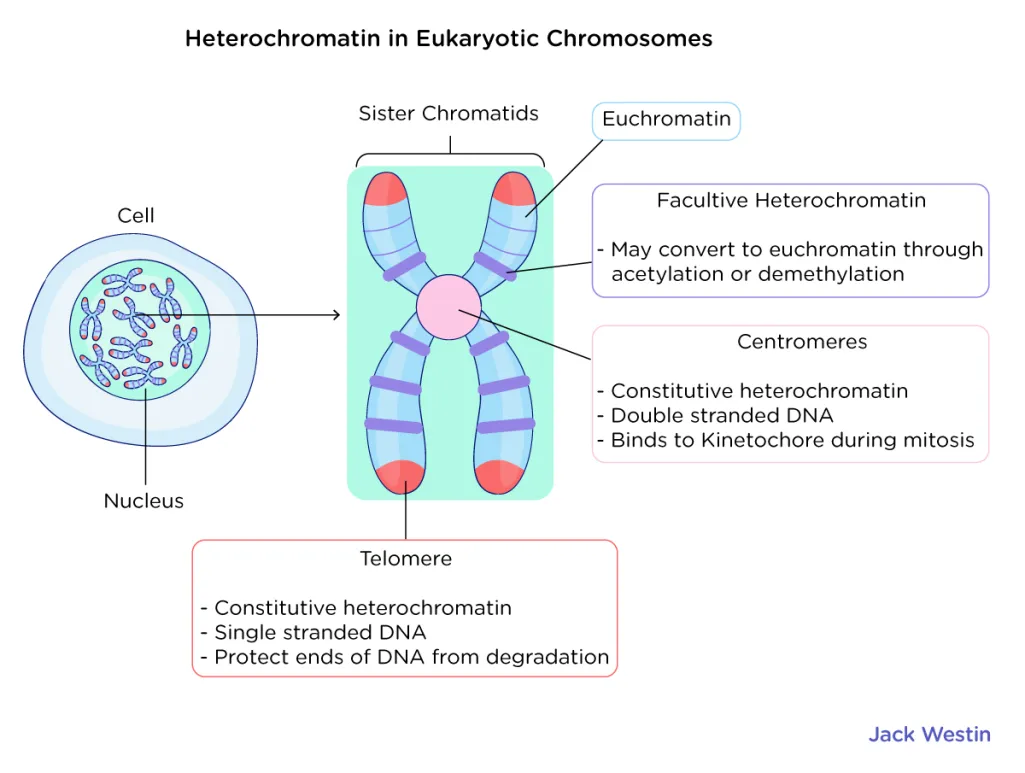
Euchromatin
-less tightly packed nucleosomes
-coincides with regular transcription
-H3—> acetylation
Acetylation of histone is linked to euchromatin (active, open chromatin).