RNA Interference and Recombination- Exam 3
1/18
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
19 Terms
steady state level of mRNA
amount of mRNA in cell available for translation
determined by combination of transcription and mRNA degradation rates
half-life of mRNA
mRNA is degraded at some point after synthesis
lifetime of mRNA varies; regulated by cell need
pathways of degradation
exoribonuclease enzymes shorten length of poly-A tail called deadenylation-dependent decay: binding of poly-A-binding protein to tail stabilizes mRNA
decapping enzymes removes 7-methylguanine cap: mRNA is now unstable
endonuclease cleaves mRNA internally
regulation of mRNA stability and degradation
adenosine-uridine rich element (ARE)
cis-acting sequence element that regulates mRNA stability
located in the 3’ untranslated region of mRNA molecules
RNA binding proteins (RBPs) bind to AREs and increase or prevent degradation
RNA interference (RNAi)
sequence specific post transcriptional regulation
short RNA molecules regulate gene expression in cytoplasm of plants, animals, and fungi; repress translation and trigger mRNA degradation
phenomena known as RNA-induced gene silencing
molecular mechanism of RNA-induced gene silencing
done by siRNA and microRNAs: short double-stranded ribonucleotides
siRNAs: arise in cell due to virus infection- produce double stranded RNA, which is recognized and cleaved by the enzyme dicer
microRNAs: noncoding RNAs that negatively regulate gene expression
siRNA pathway
double-stranded RNA is processed into siRNA by Dicer
siRNAs then associate with RISC containing an Argonaute (AGO) family protein
RISC unwinds the siRNAs into single-stranded siRNAs and cleaves mRNAs complementary to the siRNA
miRNA pathway
miRNA genes are transcribed as primary-miRNAs which are trimmed at the 5’ and 3’ ends by the nuclear enzyme Drosha to form pre-miRNAs
pre-miRNAs exported to the cytoplasm and processed by Dicer
miRNAs then associate with RISC and mRNAs
if the miRNA and mRNA are perfectly complementary, the mRNA is destroyed; if there is a partial match, translation is inhibited
non-coding RNAs
microRNAs- small noncoding fragments of RNA that regulate gene expression (thought to regulate at least 1/3 of all protein coding genes)
first discovered in C.elegans
attempts to inject long double stranded RNA molecules into worm cells was shown to disrupt translation in a sequence specific manner
is this a defense mechanism against RNA viruses
processing of siRNA and miRNA by Argonaut protein
both siRNA and miRNA are first trimmed by the Dicer protein in the cytoplasm
the smaller RNA fragments then bind to Argonaut
argonaut selects one of the two strands and then separates the unwanted strand which is then degraded
the other strand (guide strand) remains attached to Argonaut
additional proteins bind to form the RNA induced silencing complex (RISC)
guide strand recognizes a target sequence and the RISC then binds to it
RNA induced silencing complex (RISC)
double stranded micro RNAs and siRNAs are processed by enzyme dicer
dicer cleaves the RNA transcript into smaller fragments
effector complex proteins bind the cleaved double stranded RNA and make it single stranded
single strands form the RISC which binds to mRNA transcripts
if perfect match → mRNA is cleaved and degraded
if imperfect match → the RISC binds to the mRNA blocking translation
RNA-induced Transcription Silencing Complex (RITS)
specific siRNAs and miRNAs are transcribed and processed by Dicer and Argonaut and return to the nucleus
in nucleus they form the RITS
RITS binds to an RNA that is being transcribed
RITS then attracts other proteins that modify the chromatin, adding methyl groups and transforming it into heterochromatin to suppress transcription
RNAi in Biotechnology
RNAi studied in the lab
developed as a pharmaceutical agent
therapeutic RNAi attacks diseases caused by overexpression of specific gene or normal expression of abnormal gene product
potential use of RNAi in diagnosis of cancers
RNAi reduces severity of infections by viruses such as HIV, influenza and polio
successful treatment in animal models: viral infection, eye diseases, cancers, inflammatory bowel disease
immunoglobulin system in humans
antigen recognition: immune system binds to foreign substances (antigens)
humoral immunity:production of immunoglobulins (antibodies) that directly bind to antigens
B lymphocytes: B cells → four variable regions that recognize specific antigen, each B cell synthesizes only one type of immunoglobulin
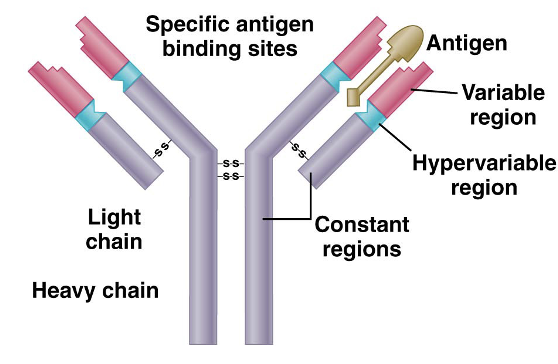
polypeptide chains immunoglobulins
consists of 4 chains
two identical: Light (L) chain and heavy (H) chain form Y-shaped structure
DNA regions organized into L(leader) and V (variable) regions
constant and variable regions
each L and H chain contains constant and variable regions: constant → C-terminus, variable → N-terminus
antibody diversity occurs in part from random recombination of 35-50 different functional LV regions with any different J (joining) regions
40 V, 5 J, and C regions recombine to form a Kappa light chain
51 V, 27 D and 6 J regions to form heavy chain
recombination and hypermutation
mechanism that further increase antibody diversity
recombination event: joins LV region to J region; not precise, results in considerable variation
high hypermutation: random somatic mutation during B cell development introduces more variation into the LVJ region’s sequence
ENCODE
encyclopedia of DNA elements project
goal: identify all functional DNA sequences and determine how elements regulate expression
discovery: more than 80% of human genome contains regulatory elements (once considered “junk DNA”)
Enhancers and promotor elements
promoters elements: 30,000 promoter regions found within protein-coding genes
ENCODE data show enhancer elements influence transcription of several different promoters
enhancer elements: contain promoter elements that initiate transcription