Lecture 10 - Epigenomics: DNA Methylation
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
21 Terms
What is epigenetics and some exmaples of expeirments?
Epigenetics
Heritable changes in gene expression that don’t involve changes in the DNA sequence
Experiment examples
Agouti mice - changing of a certain transposon can lead to darker mice
Toadflax flower - WT flower and mutant flower have identical DNA sequences
What is epigenetic information
DOES NOT CHANGE DNA SEQ
Involves:
Cytosine DNA methylation (5-methylcytosine)
Histone modifications and variants
chromatin structure and gene expression
siRNAs made by RNAi
modify chromatin
DNA methylation
ex: change transposon
All methods for altering gene expression in a non-Mendelian manner
Why do people care about DNA methylation?
Inactivates transposon and harmful DNAs
Regulate normal gene expression (ex: imprinting - diff gene expression in mother and father)
Irregular DNA methylation contributes to cancer
ex: lack of DNA methylation - proto-oncogene → oncogene
too much DNA methylation on tumor suppressor gene
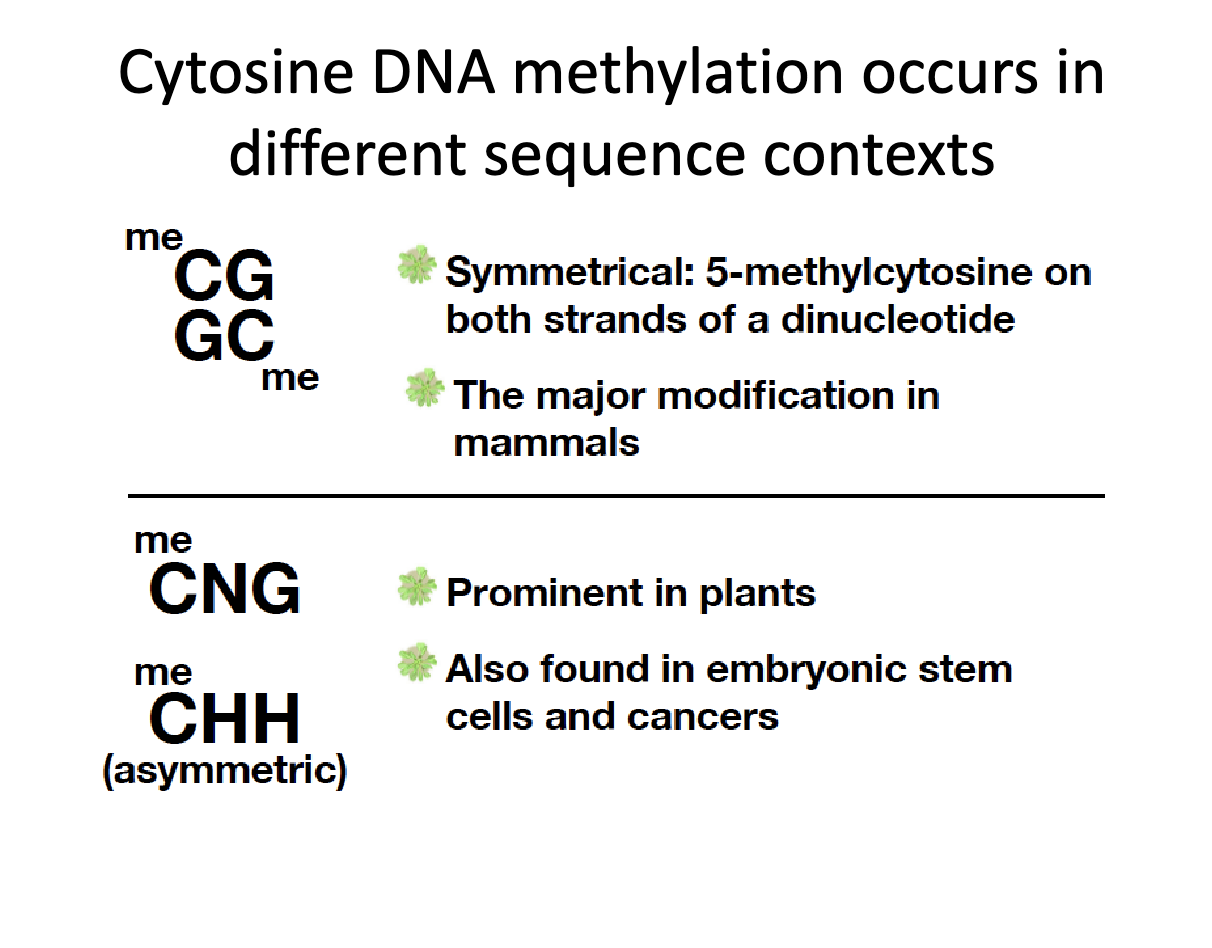
What are some different cytosine contexts?
Symmetrical
CG/GC - 5-methylcytosine on both strands of dinucleotide
major modification in mammals
Assymetrical
CNG or CHH
N = any nucleotide, H = any nucleotide other than cytosine
Establishment and Maintenance of DNA methylation
Establishment (De Novo)
DRM2 (enzyme/methyl transferase)
CG, CNG, CHH (asymmetric)
Maintenance (existing mehtylation) (ex: hemimethylated DNA)
MET1 (Dnmt1) → CG
CMT3 and DRM2 → CNG and CHH (mathylate strand of DNA that lost methyl in replication)
Relationship between DNA ,methylation and small RNAs
ex;
si RNA → H3K9me → DNA methylation
H3K9me and DNA methylation responsible for gene silencing
What is the prupsoe of Affinity Purification
anti-methyl cytosine antibodies
depends on overall density (Immunopurification subset of AP)
increased density = increase ability to purify
CH, CNG, CHH combined - MBD (methyl binding domain) doesn’t recognize diff cytosine contexts
What are the steps of Affinity Purification
1) Fragment genome with sonicator or fragmentase
2) Use antibody to recognize all methyl cytosine fragments
3) Antibody or MBD protein can take out DNA with mehtylated cytosine
4) Add Illumina adapters, SBS, barcode, P5/P7
5) seq DNA - Illumina
6) Align readsto genome
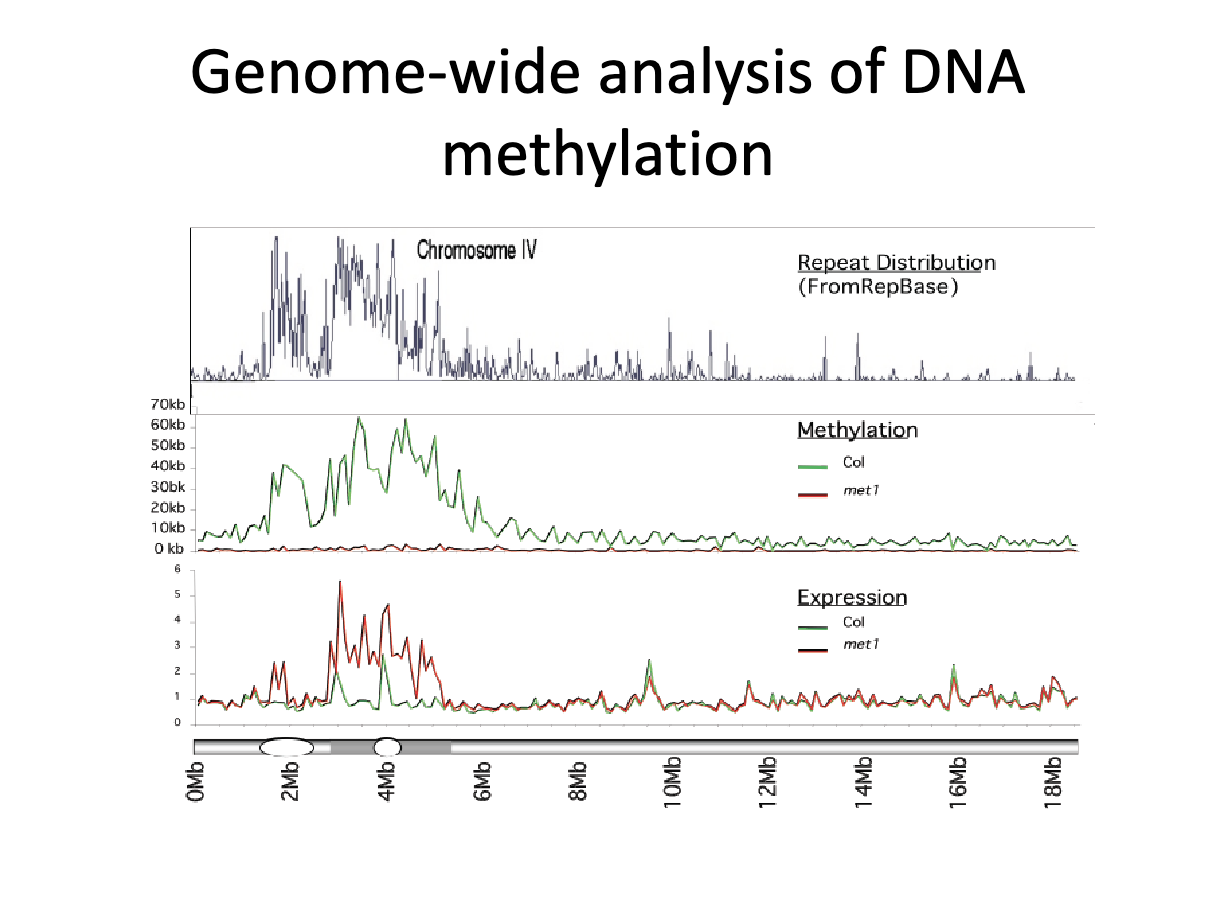
What are some takeaways from interpreting this graph?
Graph
Top graph - distribution of repeats on chromosome
middle graph - amount of methylation
bottom graph - levels of gnee expression
line at bottom - chromosome
longer oval at 2Mb - centromere
smaller oval at 4 Mb - heterochromatic knob - increased heterochromatin density
Takeaways
Areas of high repeat density overlap with centromere
Areas with high repeats have high methylation
Areas of DNA methylation no expressed strongly
Without met1 gene methylation is lost, but cytosine context now known since affinity purification used
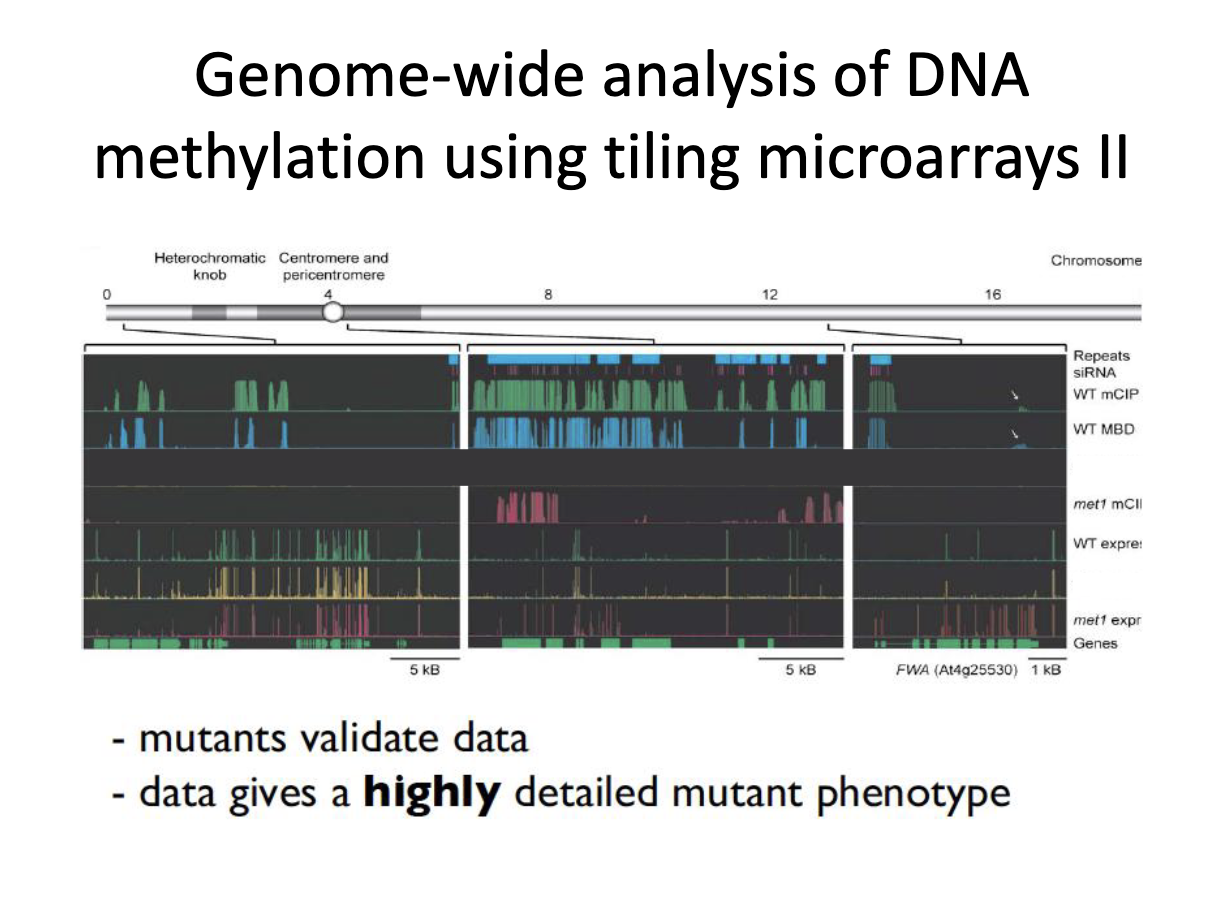
What are takeaways form this graph?
Takeaways
areas of methylated cytosine correlate with regions of repetitive regions and siRNA (but not always)
If you lose met1 gene, lose majority of methylation
Methylation slight correlates with the repression of gene expression
Mutants validate data and data gives highly detailed mutant phenotype
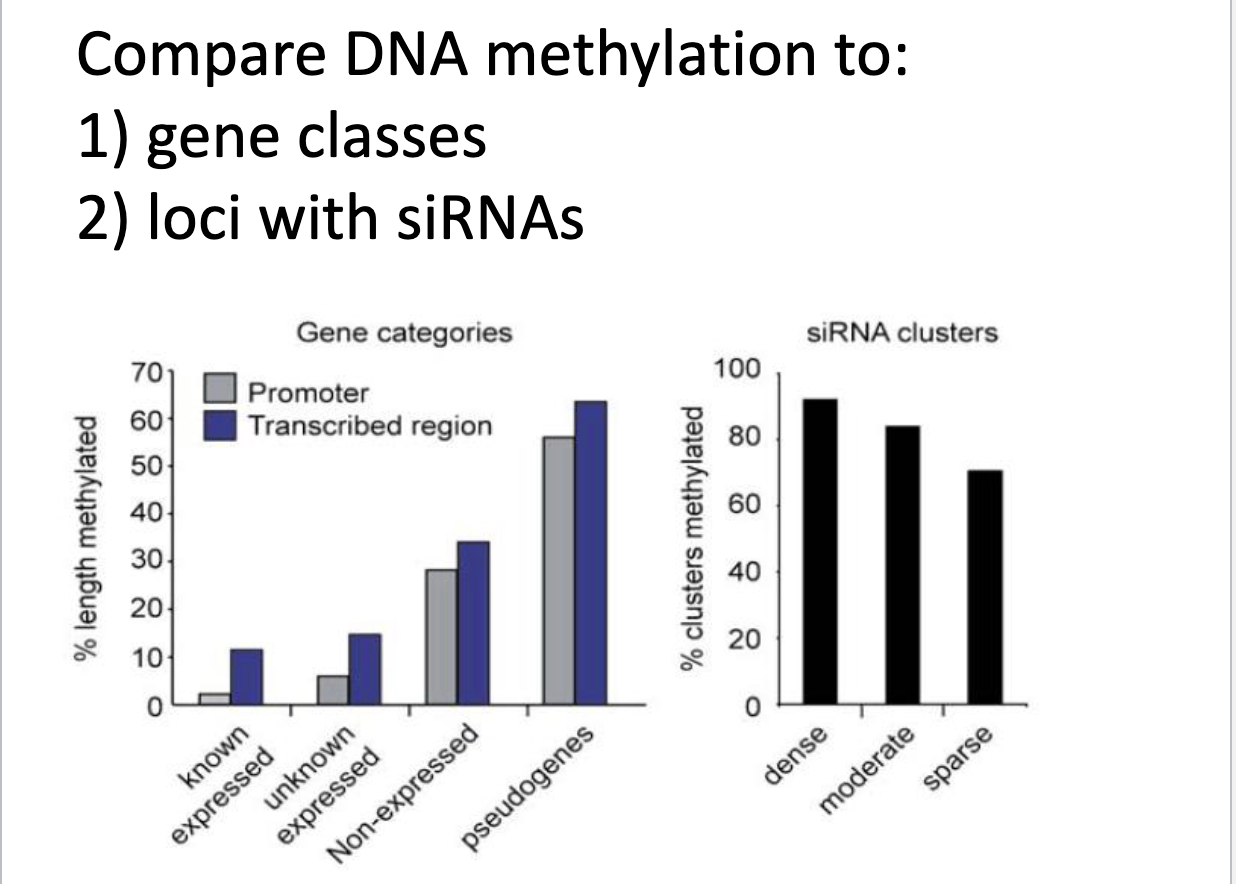
What can you take away from these bar graphs
Graph 1
x-axis: different gene categories; y-axis: relative length methylated
Graph 1 Takeaways
Shows genes that are known to be methylated, not known to be methylated, not expressed, or non functional (psuedo)
Less methylation is promoters than in transcribed genes regions
Expressed genes are less methylated than psuedogenes
Graph 2
y-axis: relative clusters methylated; x-axis: siRNA clusters of varying densities
Graph 2 Takeaways
the denser the siRNA clusters are, the more methylated they are
Describe the steps of Sodium bisulfate sequencing
1) Denature the genomic DNA since methylated cytosine can be protected in doulbe strand form
2) Treat the denatured DNA with Sodium bisulfite
3) Fragment DNA
4) add Illumina adapters
5) Amplification → get DNA copies of SBS treated DNA
6) P5 + P7 oligos
7) Illumina sequencing
8 ) Align to genome
Look fors cases where:
C is now a T → cytosine was NOT methylated
C is still a C → cytosine WAS methylated
Affinity purification with an antibody to methyl cytosine coupled with sequencing is able to determine cytosine context. True or false.
False.
Sodium bisulfite sequencing is able to determine cytosine context. True or false?
True
DNA methylation is ONLY associated with repetitive regions and regions from where siRNA arise. True or false?
False → can also be in other genomic regions
Denaturing double stranded DNA ensures that the antibody to methylated cytosine or the sodium bisulfite can access all unmethylated cytosines. True or false?
True → double stand can protect methylated cytosine
Two phenotypes of a mutant in MET1 are reduced DNA methylation and increased gene expression. True or false?
True
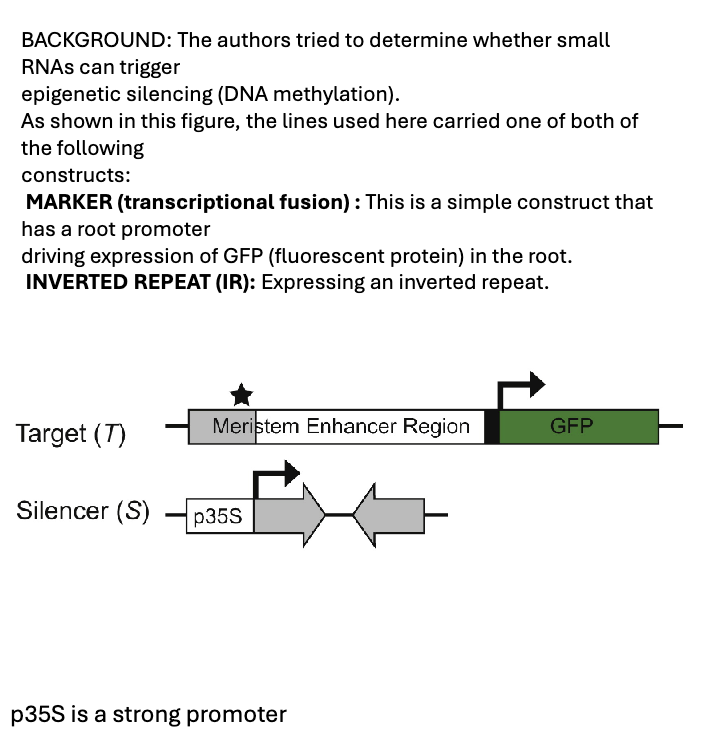
1. As shown in this figure, the inverted repeat in the IR construct will generate an RNA that can fold onto itself (no bulges). What will this trigger?
A. The production of siRNAs
B. The production of miRNA
2. One portion of the inverted repeat sequence in the IR is complementary to the root promoter of MARKER (areas shown in grey). What will the siRNAs
target?
A. Only the IR construct
B. Only the Root MARKER
C. Any sequence that is similar to the inverted repeat, including the IR and the MARKER Promoter
3. Based on what we know about RdDM, what will these siRNAs trigger?
A. DNA methylation at the target regions
B. Production of miRNAs at the target regions
C. The expression of GFP
1) A. The production of siRNAs
2) C. Any sequence that is similar to the inverted repeat, including the IR and the MARKER Promoter → siRNA target any complementary sequences
3) A. DNA methylation at the target regions → siRNA involved in DNA methylation
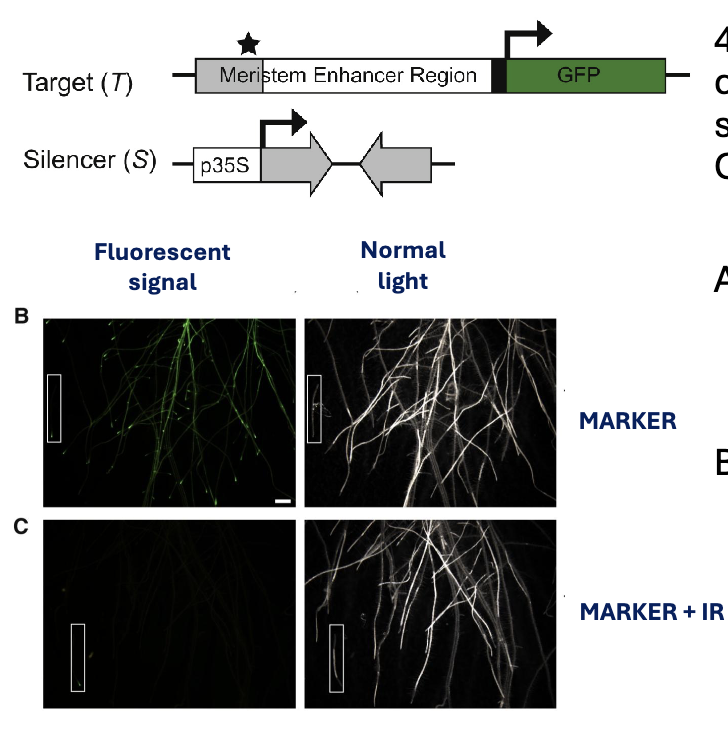
4. Do both plants in figures B and C carry the MARKER construct that
should express GFP? Why is the GFP signal not present in figure C?
A. Yes, because the plant in figure C also carries the IR construct, that silences the MARKER construct
B. No, because the plant in figure C only carries the IR construct.
A. Yes, because the plant in figure C also carries the IR construct, that silences the MARKER construct
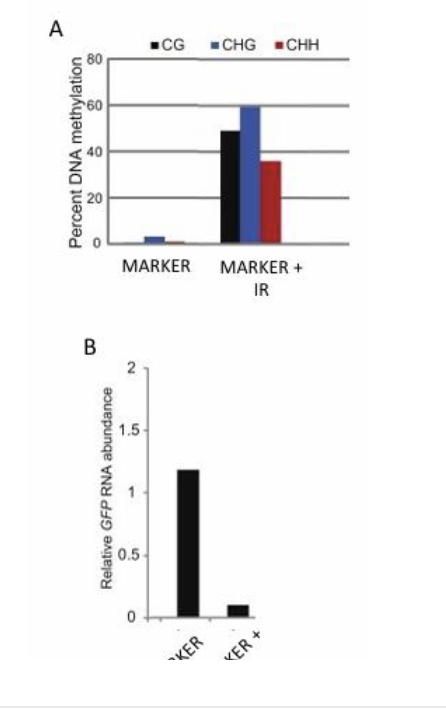
5. The authors determined methylation status (CG, CHG and CHH) in these lines. What method did they most likely use?
A. Sodium bisulfite sequencing
B. Affinity purification with anti-methylcytosine antibodies
C. RNA sequencing
D. Small RNA sequencing
6. In graph “B”, what do the x-axis and y-axis represent?
A. X-axis = Root transgenic lines; Y-axis = The GFP
B. X-axis = Root transgenic lines; Y-axis = The methylation
7. Based on “A” and “B”, how does the level of DNA methylation in a gene correlate with the abundance of its transcript? Positively or negatively?
A. Positively. The more methylation in the promoter, the higher the expression of the transcript
B. Negatively. The more methylation in the promoter, the lower the expression of the transcript.
5) A. Sodium bisulfite sequencing → only technique that can find context
6) A. X-axis = Root transgenic lines; Y-axis = The
GFP
7) B. Negatively. The more methylation in the promoter, the lower the expression of the transcript.
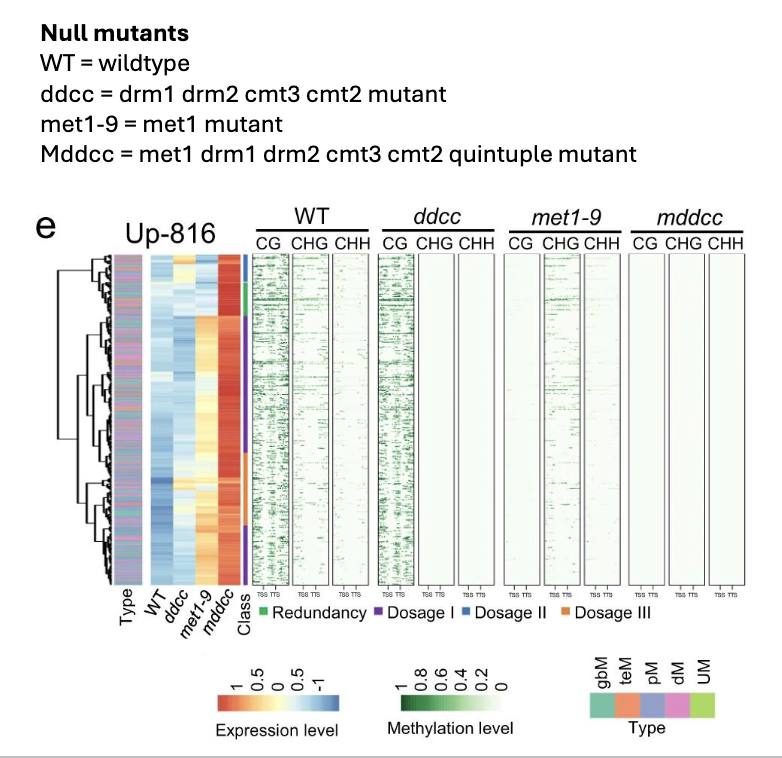
Graph context: null mutants created to determine protein’s role in cytosine methylation in Arabidopsis
8. Based on this plot which type of methylation is met1 primarily responsible for?
A. CG
B. CHG
C. CHH
D. CHH and CHG
9. Based on this plot which type of methylation is drm1 drm2 cmt3 cmt2 primarily responsible for?
A. CG
B. CHG
C. CHH
D. CHH and CHG
10. Based on this plot whicht type of methylation is plays the largest role in gene
silencing in Arabidopsis?
A. CG
B. CHG
C. CHH
D. CHH and CHG
8 ) A. CG
9) D. CHH and CHG
10) A. CG