CRIPSR, cancer, polyploidy, gene dosage, aneuploidy
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
48 Terms
forward genetics answers.. what is the process?
what genes are involved in biological process + function?
mutagenenisis screen
chemical induces mutation (parent)
self fertilized progeny (may be induced
pros and cons of forward genetics
p: easy to create new mutations
c: no specitivity, hard to determine which gene is mutated, need to look thru 100s to find phenotype
reverse genetics- what does it find out, what is the process?
what biological processes does a gene play a role in + function of gene
targeted mutagenenisis
introduce plasmid with mutant sequence
recombination, gene is in chromosome
pros and cons of reverse genetics
p: specific, discovers new functions
c: challenging to synthesize DNA seq
What was observed in Duchenne muscular dystrophy
small deletion (one nucleotide), reading frame changed, early stop codon= severe phenotype, protein truncated, LOF
large deletion (multiple of 3) but reading frame intact: onset, much less severe phenotype
when exon 23 cut out, restored reading frame= mild symptoms
oncogene
mutated gene, presence stimulates cancer GOF, derived from proto-oncogene which regulates cell growth
1 mutated copy (+ / -) = dominant negative and haploinsufficient
tumor supressor gene
absence promotes cancer
2 mutated copies (- / -) = lsot of inhibitory factors, uncontrolled cellular division
loss of heterozygosity
inactivation of functioning allele in heterozygotes, so only mutant is present
translocations on chromosomes can cause cancer promoting mutations, how? reciprical translocation
translocated next to enhancer for B cells, hella antibodies, + extra abilities
philadelphia chromosome (BCR/ABL fusion protein)
makes a hybrid oncogene/ fusion gene, half of one and another, new function makes cancer (BCR/ABL fusion protein)= overactive kinase that promotes uncotnrolled cell division, expressed in WBC
euploid, diploid, polylpoid
n, 2n, 3n4n5n…
why is polyploidy more common in plants than animals
can survive but result in sterility, interferes with meiosis
why are polyploidy tissues and plants larger
increased number of chromosomes, increases the amount of DNA in the nucleus, which often causes the nucleus and overall cell to enlarge + higher gene dosage can lead to higher production of proteins and enzymes,
aneuploidy (trisomy) vs polyploidyl (triploidy) in humans
Trisomy involves an extra copy of one specific chromosome (47 total),
triploidy involves an entire extra set of chromosomes (69 total).
infertile because chromsomes can’t divide evenly during meiosis
autopolyploidy
has 2+ sets of homologous chromosomes, all derived from same species
allopolyploid
polyploid with multiple sets of chromosomes that were derived from different species (hybridization)
two diploid species make hybrid, sterile because chromosomes cant pair properly,
if hybrid produces unreduced gametes (2n=10) = restores fertility
euploid
correct number of chromosomes
haploid n =23
diploid 2n=46
polyploid banana 3n=33, whole number multiple of base set (n=11)
polyploidy and reproductive isolation
polyploids can’t produce viable with others offspring due to mismatched chromosome numbers
polyploids can only breed with others of same ploidy level = reproductive isolation and speciation
comapred to XY individuals, XX individuals would have twice the dosage of X linked genes, hwo is this corrected?
XCI lionization: during embryogeneisis, one X chromosome becomes barr body (inactivated chromsome, heterochromatin), ensures proper dosage so X and Y have equal expression of X linked genes
mosaic cell: cell that is geneticlaly different from other cells in same individual
occurs in XX individuals due to random X chrosomosome inactivation, XX people are mosaics for X-linked gene expression- mix of normal adn affected cells
mutation types from DSB in different parts of a gene
promoter/ regulatory:
start codon:
early coding sequence (exon)
middle of coding region
end of gene, stop codon (last exon)
promoter/ regulatory: reduce/ stop transcription (LOF)
start codon: no translation (LOF)
early coding sequence (exon): frameshift, nonsense (LOF)
middle of coding region: missense, delete protein segments (partial LOF)
end of gene, stop codon (last exon) protien mostly fucntional (mild LOF/ no major effect)
mutation types depending on repair pathway
NHEJ:
HDR:
NHEJ: DNA ends joined directly, with errors- indels, frameshift, gene knockout LOF
HDR: use of provided template to repair break, percise edit, gene correction, GOF
if there's a deletion in the UTR does that cause frameshift
no! frameshift mutations occur in coding regions, so will not directly affect AA sequence, but can impact gene expression
5’ UTR: can affect ribosome binding for translation initiation, regulatory elements- LOF
3’ UTR: determines cleavage and polyadenylation, mRNA stability- LOF
give an exmaple of mosiacism with anhydrotic dysplasia
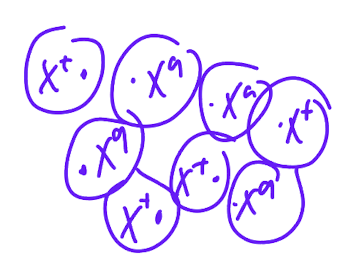
When X+X+ crossed with XaY, girls are XaX+, in embryogeneisis (early mitosis) random inactivation (barr body)= msoaic expression in cell populations of females, some express normal, other expresses mutant,
once X is inactivated in cell, daughter cells maintain inactivation pattern
phenotypically: if cirtical number of genes have mutant, sweatglands obersvable
barr body formation
before eymbryogensis
transcription from one X chromosome per cell
long non coding RNA (lnRNA) premotes heterochromatin and silencing
Whichever X chrom expresses Xist first becomes Barr Body
why does gene dosage matter
leads to imbalanced pathways- too much promoter/ inhibtion
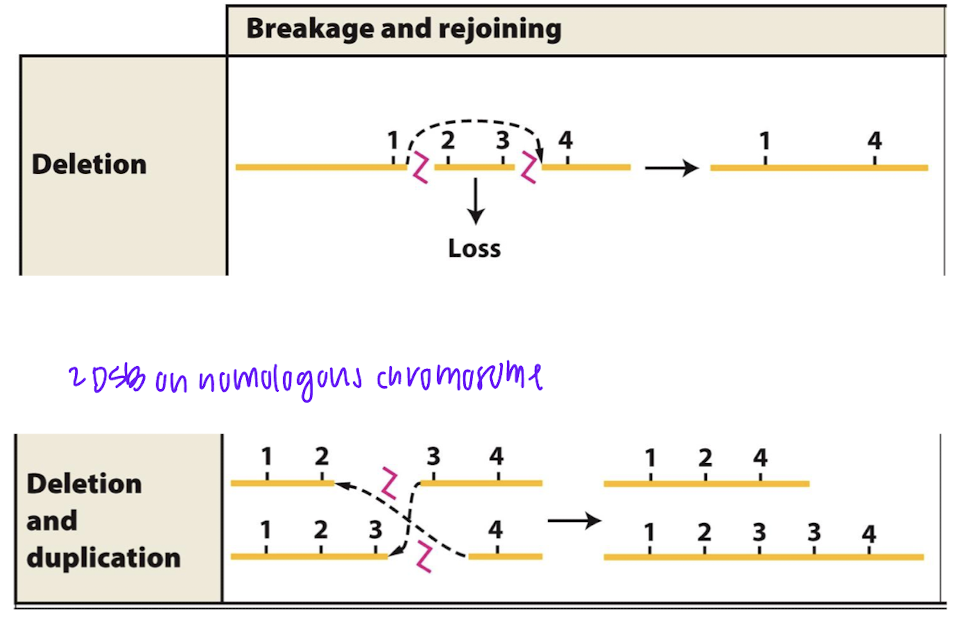
how are chromosome deletions and duplications formed
highly repetitive sequence
crossover event
if sisters are imporperly aligned with sequences, can lead to deletion and duplication
more seen in x chromosomes
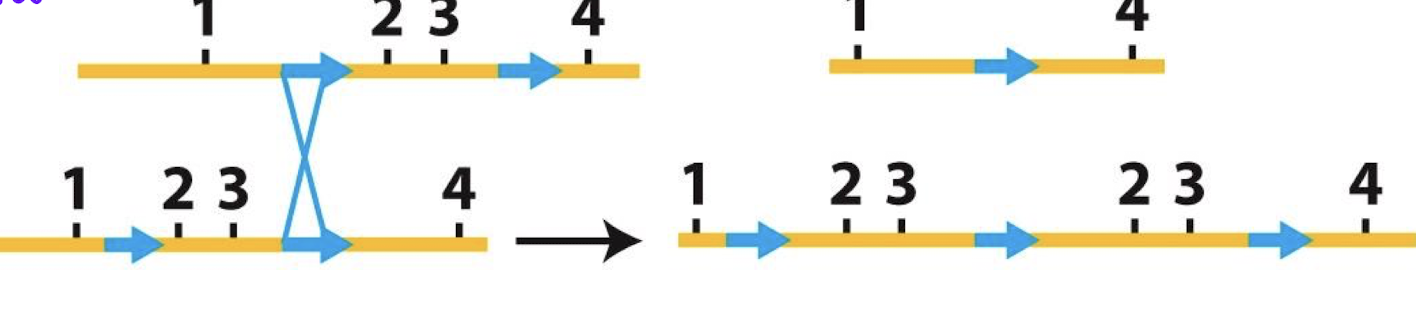
paracentric
inversion (flipped sequence DEF, FED) does not include centromere
paricentric
inversion includes centromeret
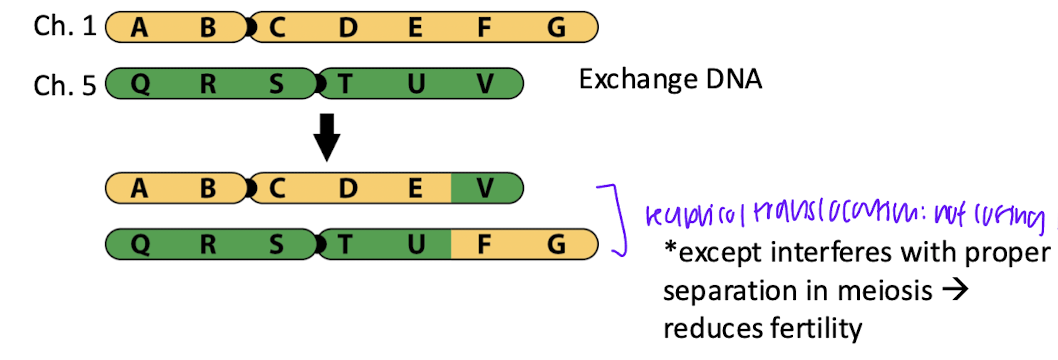
reciprocol translocation:
transfer of DNA between non homologous chromosomes
not losing info, just moved t o new chromosome, oftenWT phenotype because not disrupted gene dosage
inversion loops at meiosis- pericentric inversion
homologous chromosome: help inverted chromosomes to seperate by paring wiht homolgous regions
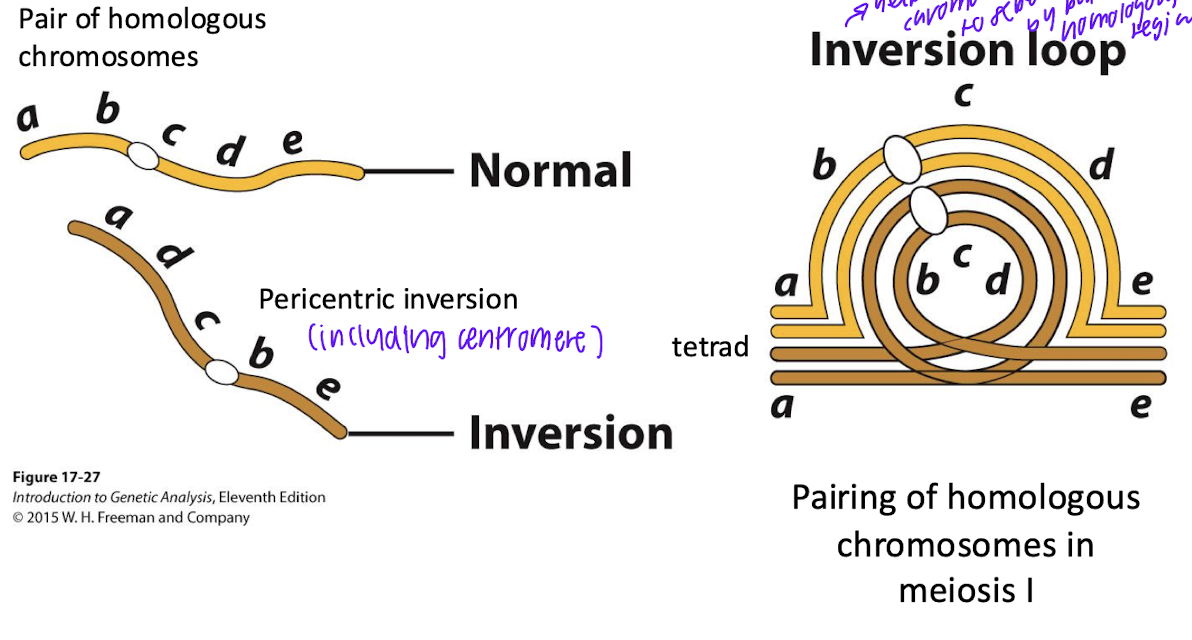
paracentric inversions can lead to deletion products. how?
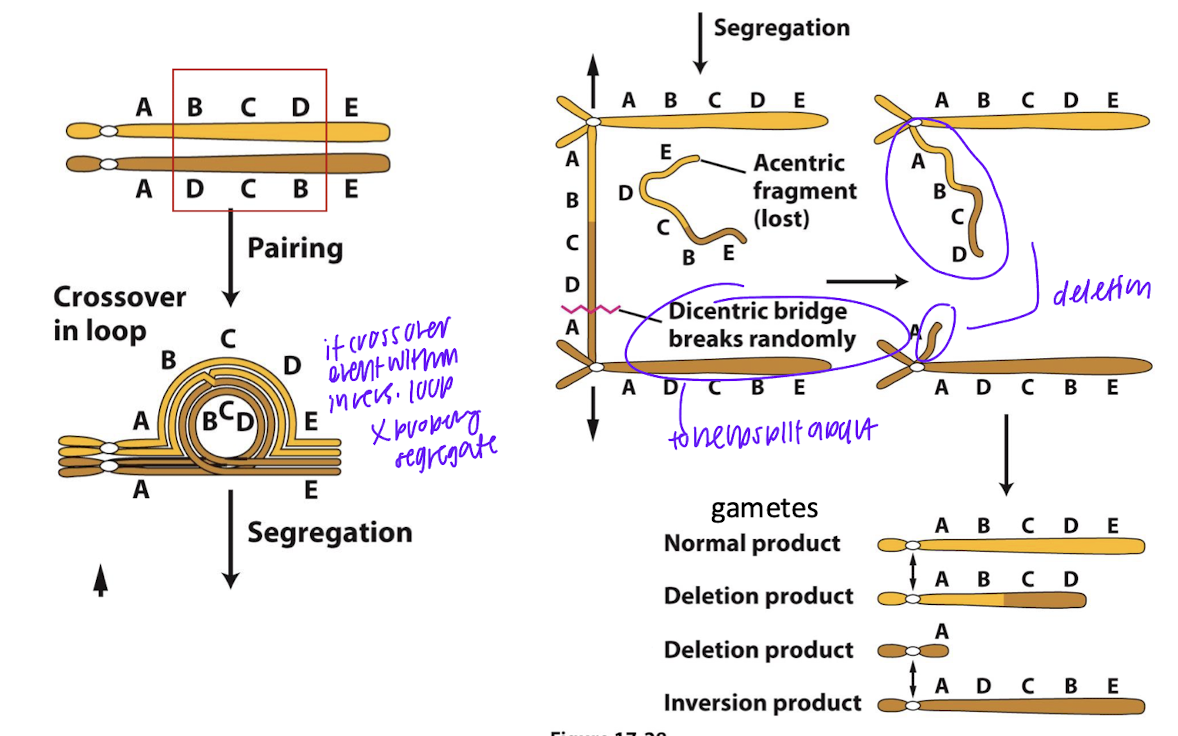
homo chromosomes, if crossover event within inversion loop doesnt properly segretate, dicentric bridge breaks randoly, acentric fragments get deleted
how does reciprovcal translocation happen?
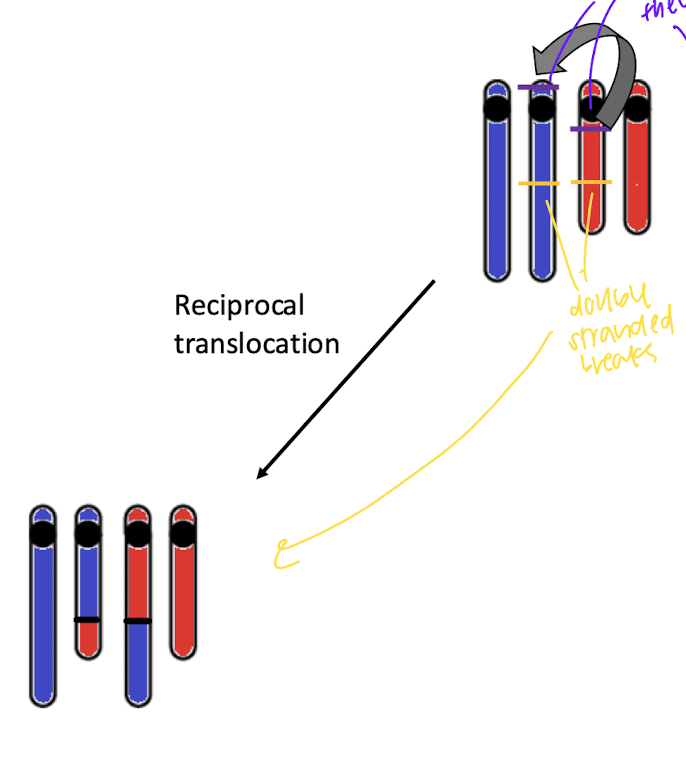
breaks near centromere and double stranded breaks on chromsome
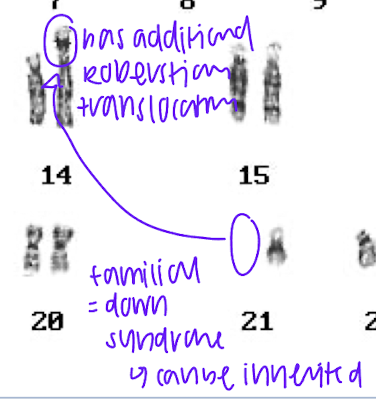
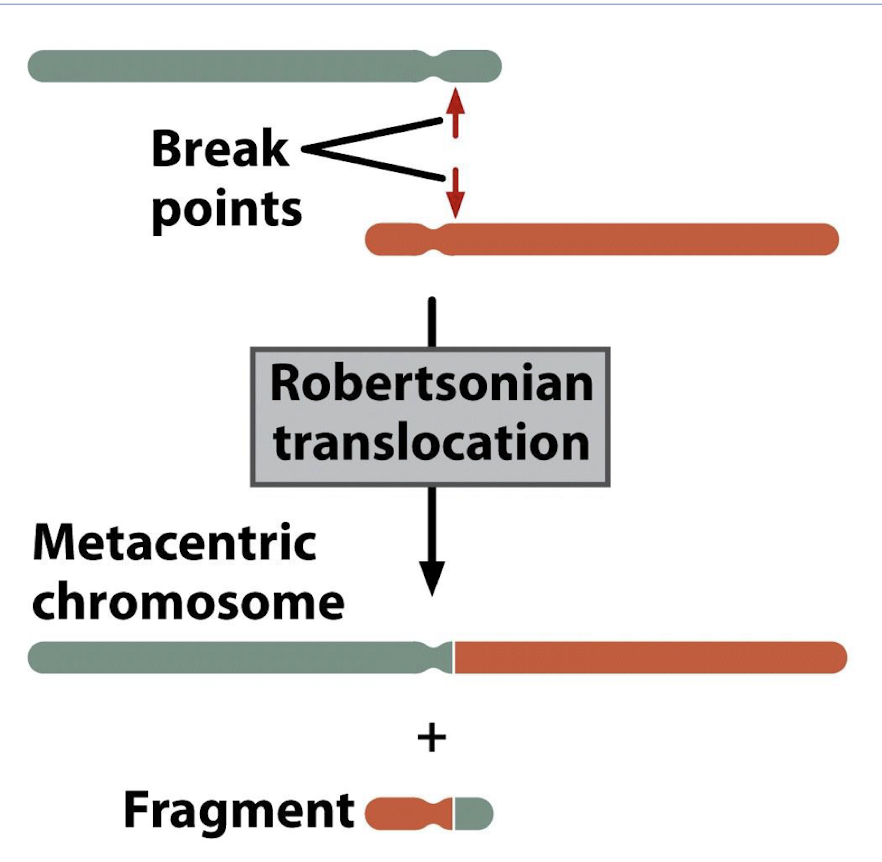
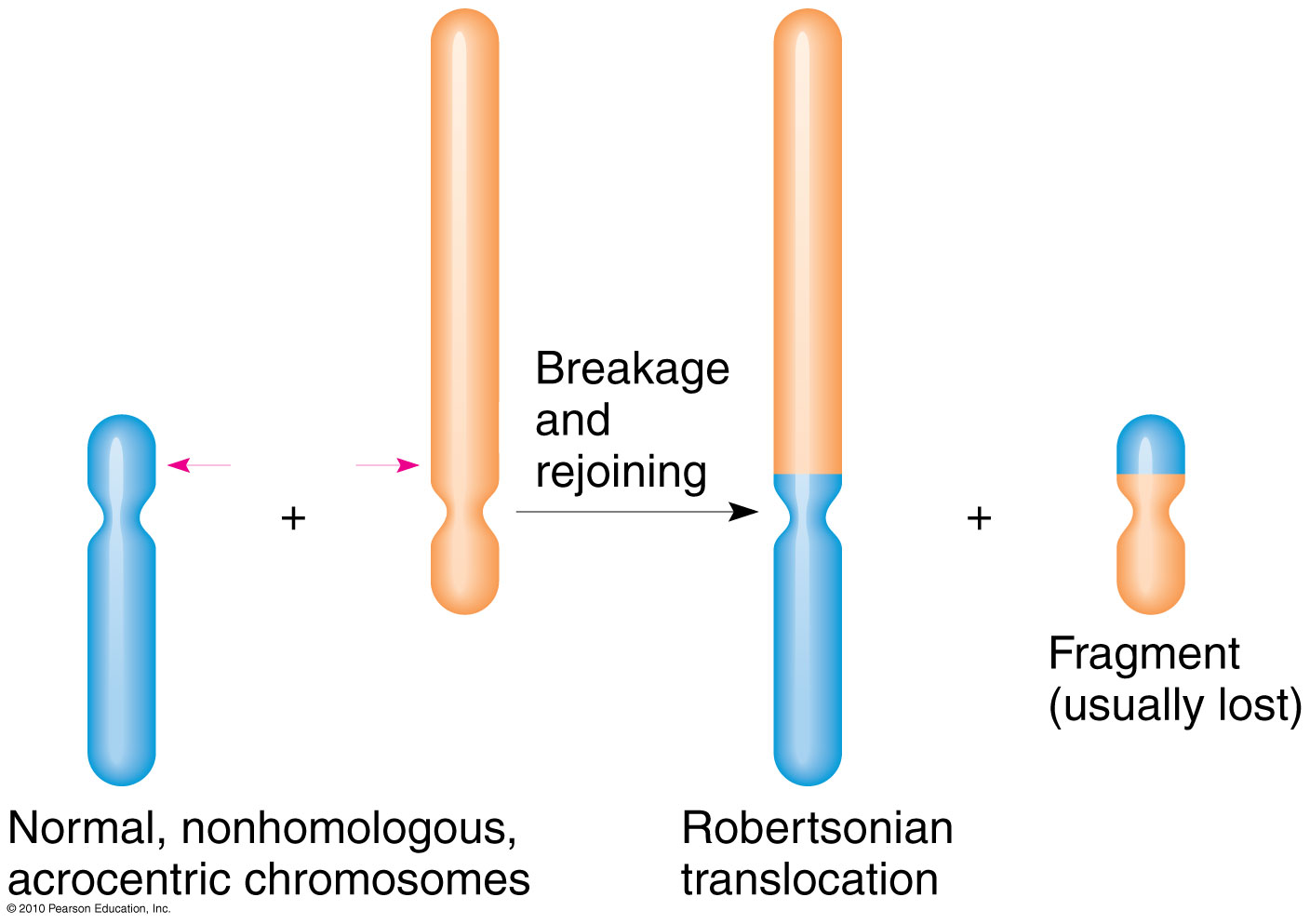
how does robertsonian translocation happen, what does it look like on the karyotope, what is the disease associated with this
acrocentric chromosome- centromere is located near one end, resulting in a very short arm (p) and a long arm (q)
break near centromeres, form metacentric chromsomes, small fragment is lost
genes are now in different location, BUT GENE DOSAGE IS THE SAME
,
what are the gametes from robertsoain translation
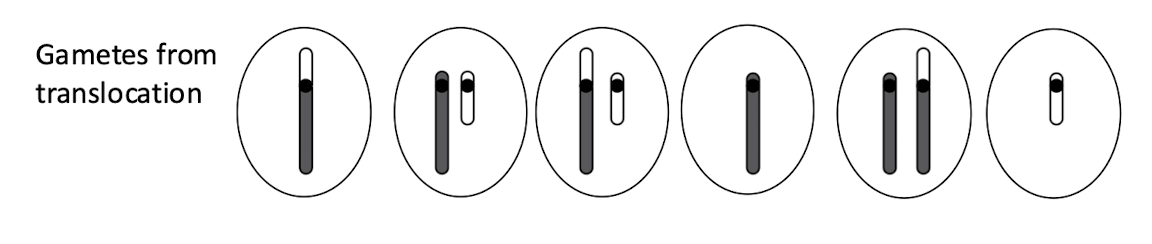
what is down sydrome caused by
tirsomy 21
why are almost all trisomies and monosomies lethal- except x chromosome
trisomy: bar body, only expression of one x chromosome, neutralizes
monosomy: only one X is expressed anyway
why does risk for down syndrome and other trisomies increase by maternal age
a phase that begins in the fetal ovary and is only completed decades later, during ovulation
cohesions can weaken
spindle fibers that properly segerate chromosomes in meiosis
do inversions result in change in gene dosage
no
role of NAHR and repetitive sequences
non-allelic homologous recombination sequences that are not from same location on chromosome, share high degree of similarity=
repeitative DNA sequences misalign leading to unequal crossing over
deletions
phenotypic effects
meitotic effects
evolutionary implications
strong effect ebcause loss of dosage can lead to haploinsufficiency, foten deleterious and selected againstdupl
duplications
increase doasage= may alter gene regulation, major soruce of new genetic material
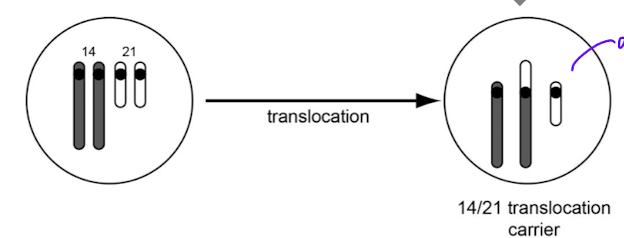
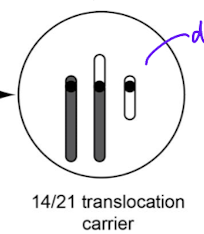
difference between sporadic and familial down syndrome
sparadic: trisomy 21 due to non disjunction in meisosis, not inerhited
familial: due to translation of chromosome 21 onto chromosome 14, can be passed down
hallmark characteristics of cancer
constant proliferation, no growth supressors, resisting cell death, activating telomerase for immortalility
insertions happen when
DNA poly slips on newly synthesized strand, the second round of replication is used as template and bp are inserted
slippage happens when
mononucletie, dinucletodie, tadem repeats
how must mutations happen?
second round of replication- incorporated into DNA
non reciprocal
non-homologous chromosomes: one chromosome gaining genetic material while the other loses it, loss in gene dosage
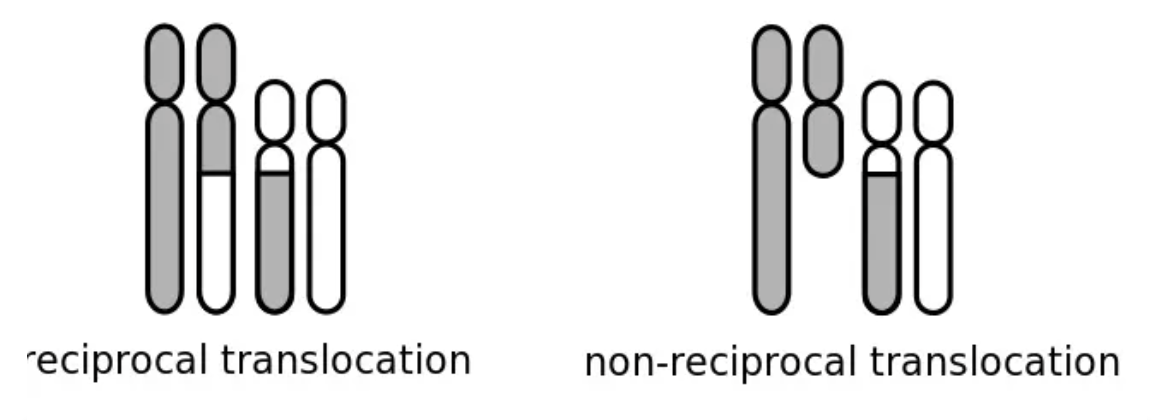
two ways translocation can make mutation
change of expression
can relocate to new area near enhancer
fusion gene
can make hybrid