VM 602 Physiology
1/510
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
511 Terms
why do we need proteins?
provide structural support
function as enzymes
involved in signaling
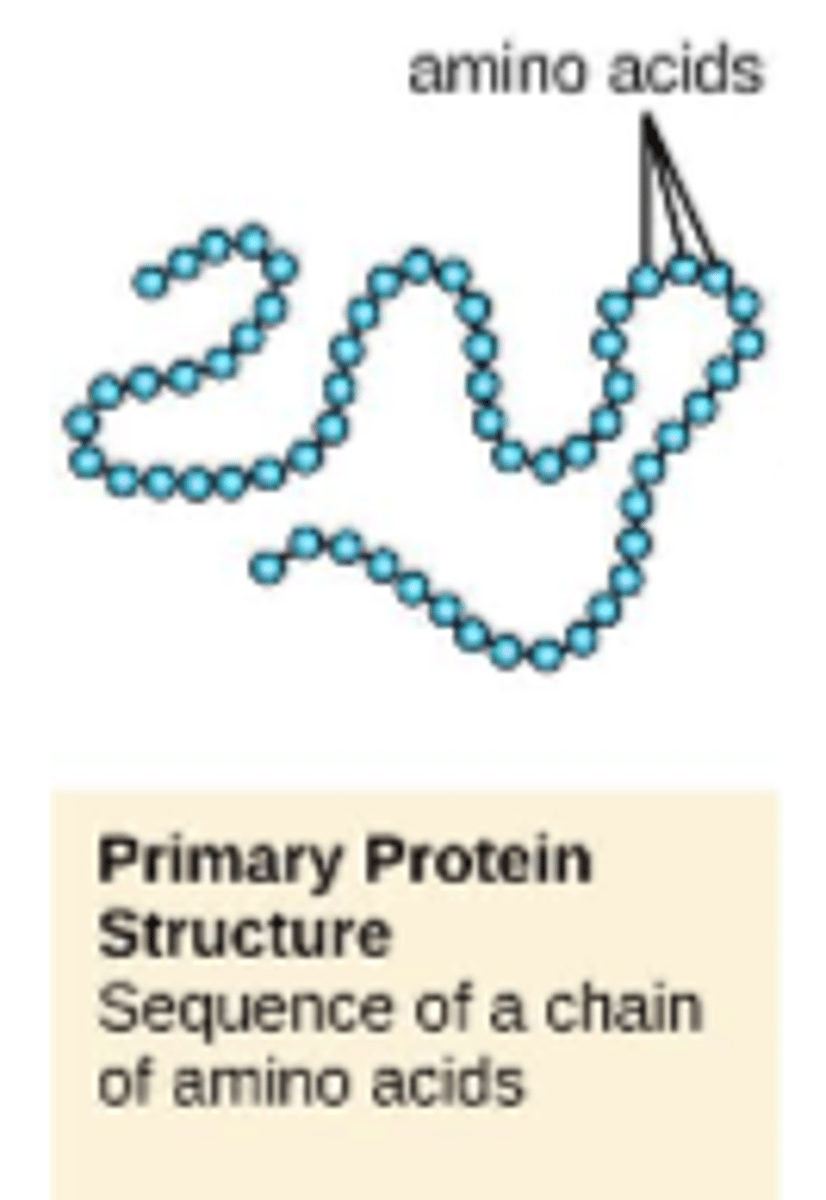
proteins are made up of
amino acids
amino acids are linked together by _______ _________ bonds
covalent peptide
multiple peptides link together to form
polypeptide backbone
4 levels of structure of protein
primary, secondary, tertiary, quaternary
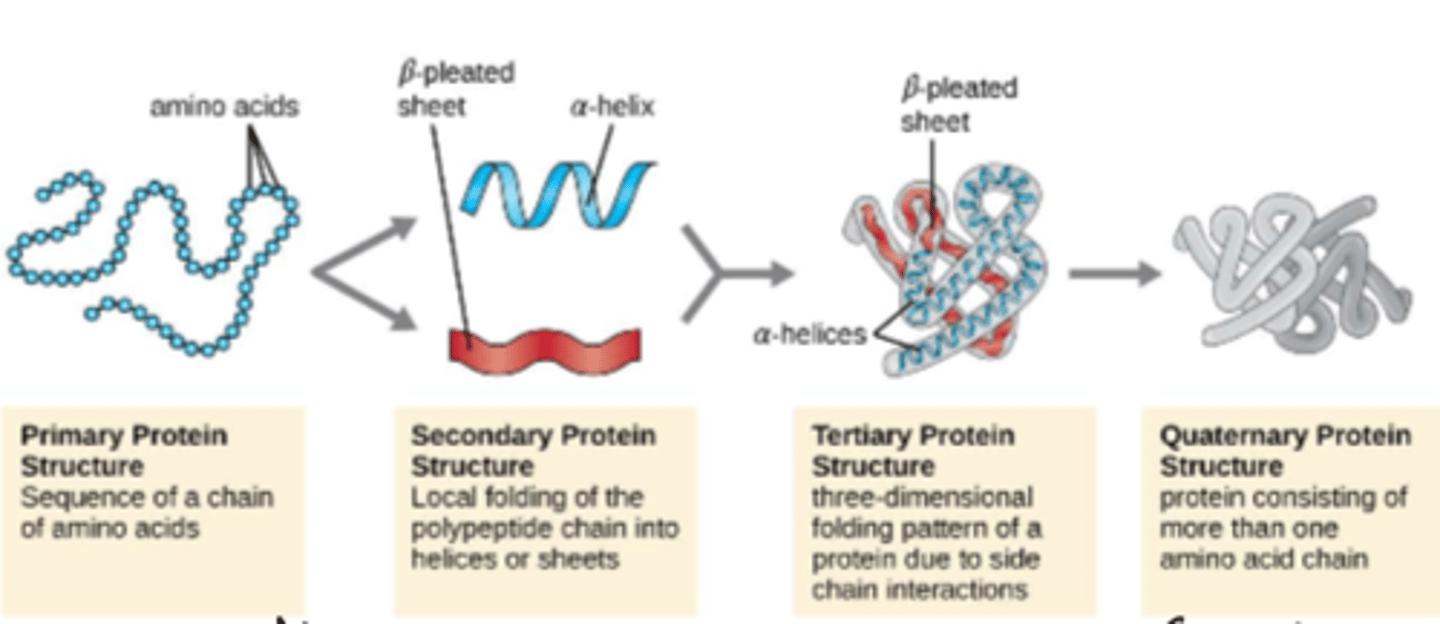
proteins are made up of amino acids written from N- to C- terminus
primary structure
a regular, repeating conformation in a protein
-includes a-helices, B-sheets
secondary structure
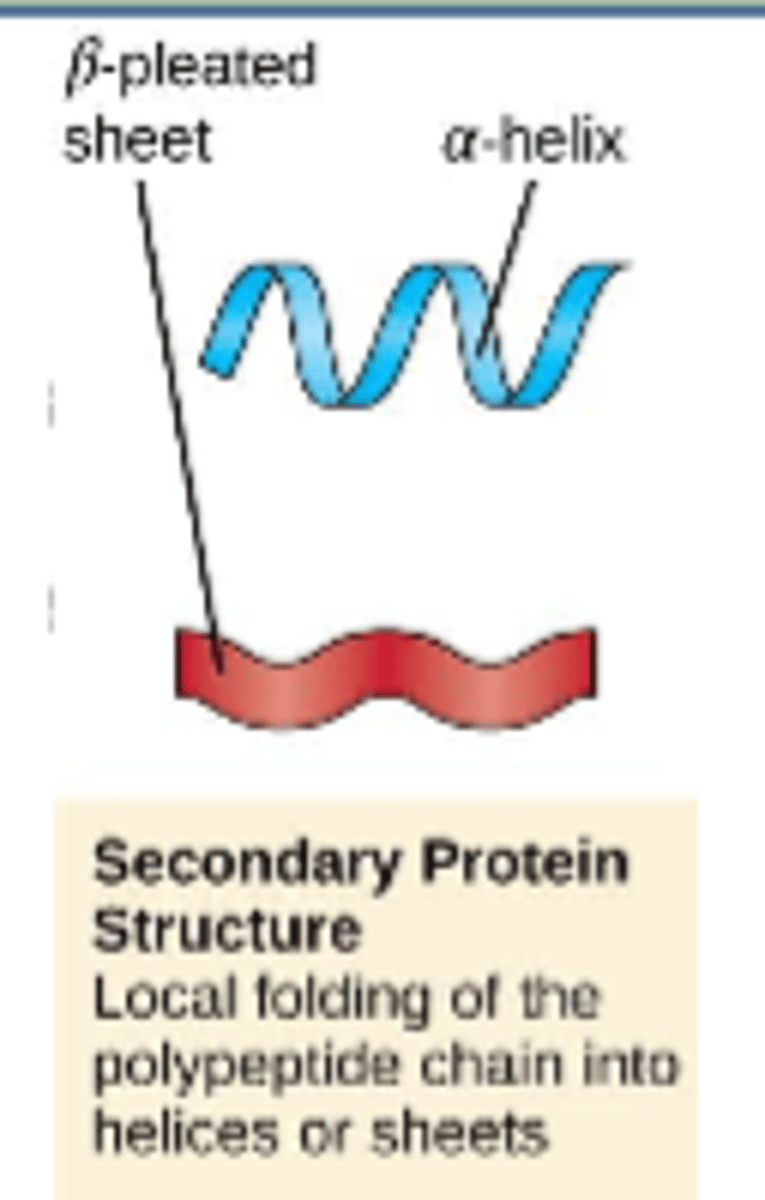
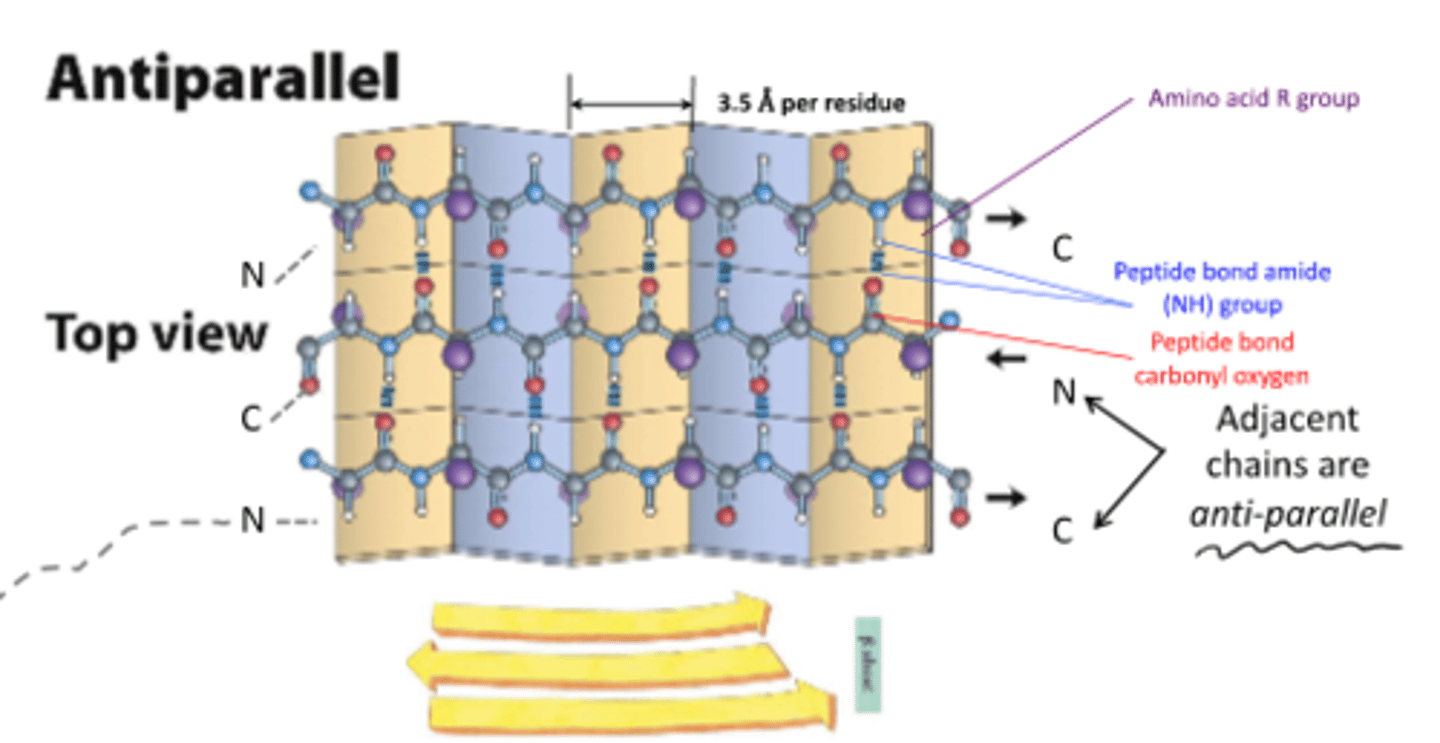
secondary structures found in proteins are a-helices and B-sheets. both have regular repeating patterns that maximize:
hydrogen bonding
adjacent chains in B-sheets are
anti-parallel
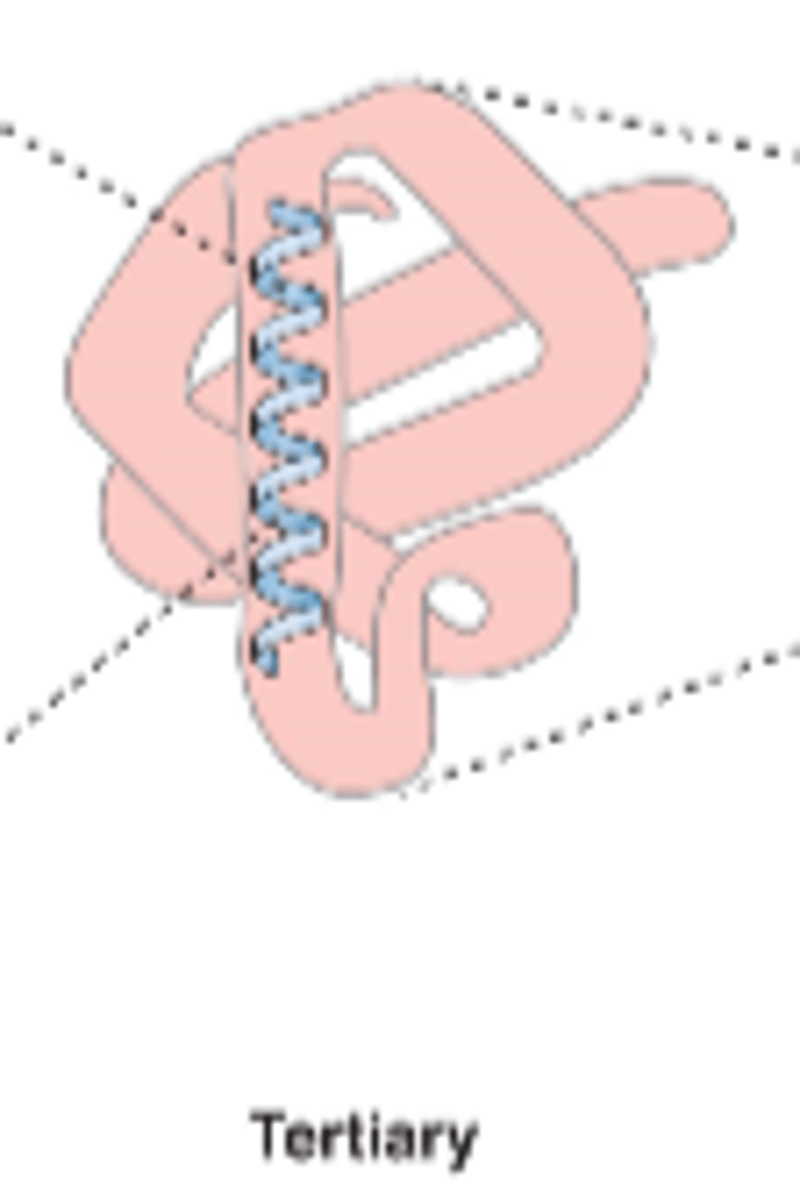
the overall 3D arrangement of amino acid residues
-3D folding of a single polypeptide chain
tertiary structure
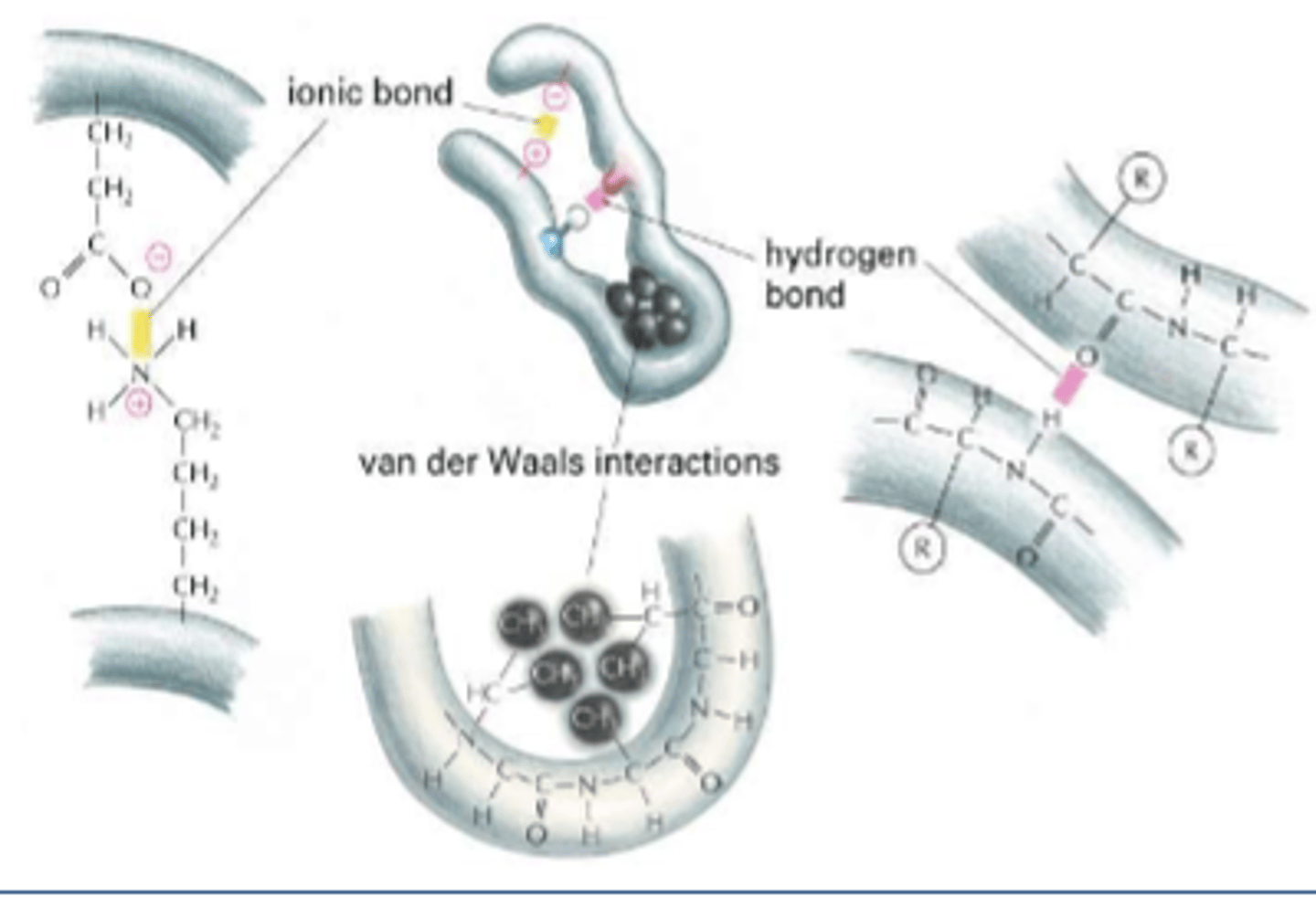
tertiary structure bonds
H bonds
disulfide bonds
ionic bonds
van der Waals interactions
hydrophobic interactions
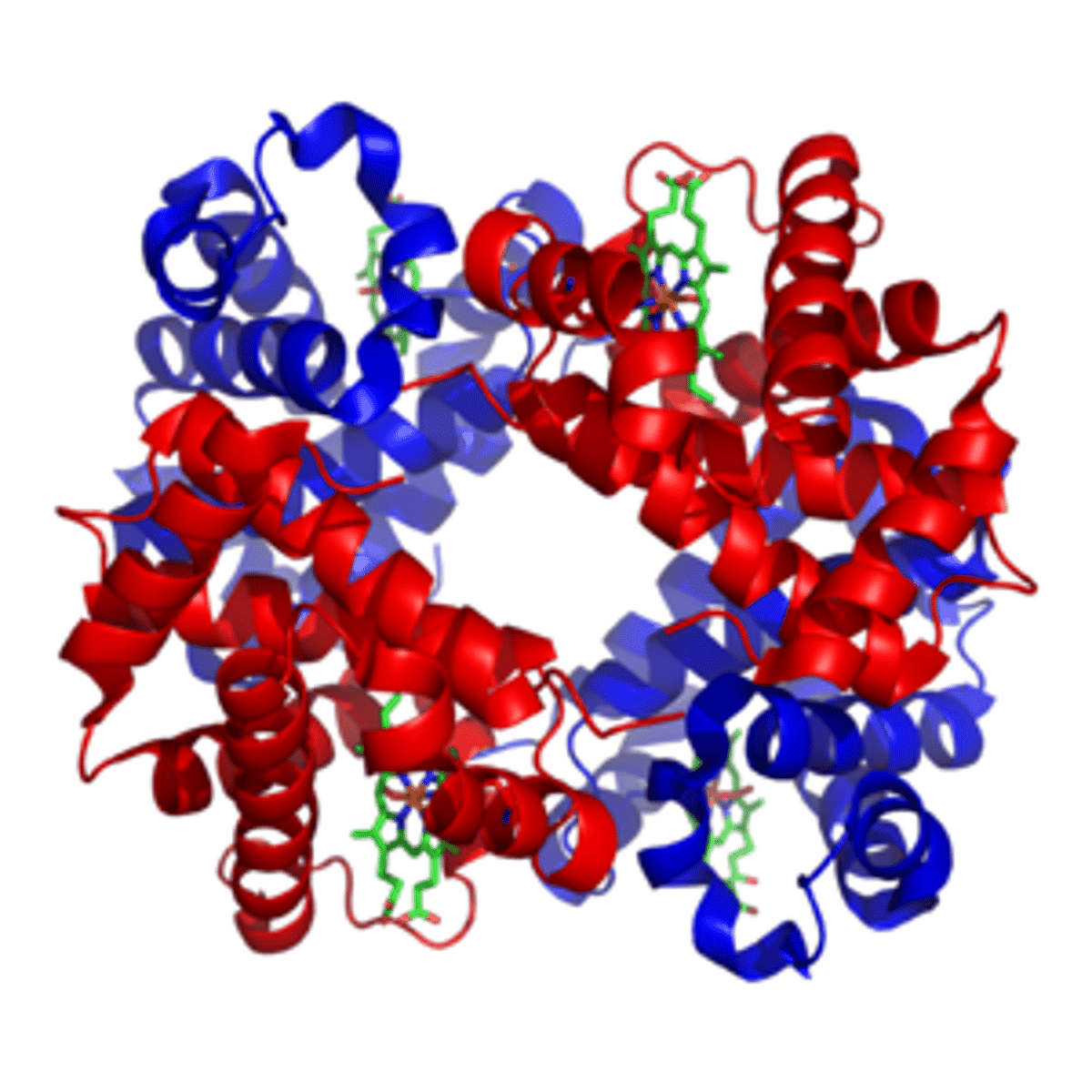
for multi-subunit complexes, this is the 3D arrangement of the subunits (multiple polypeptide chains with multiple subunits)
quaternary structure
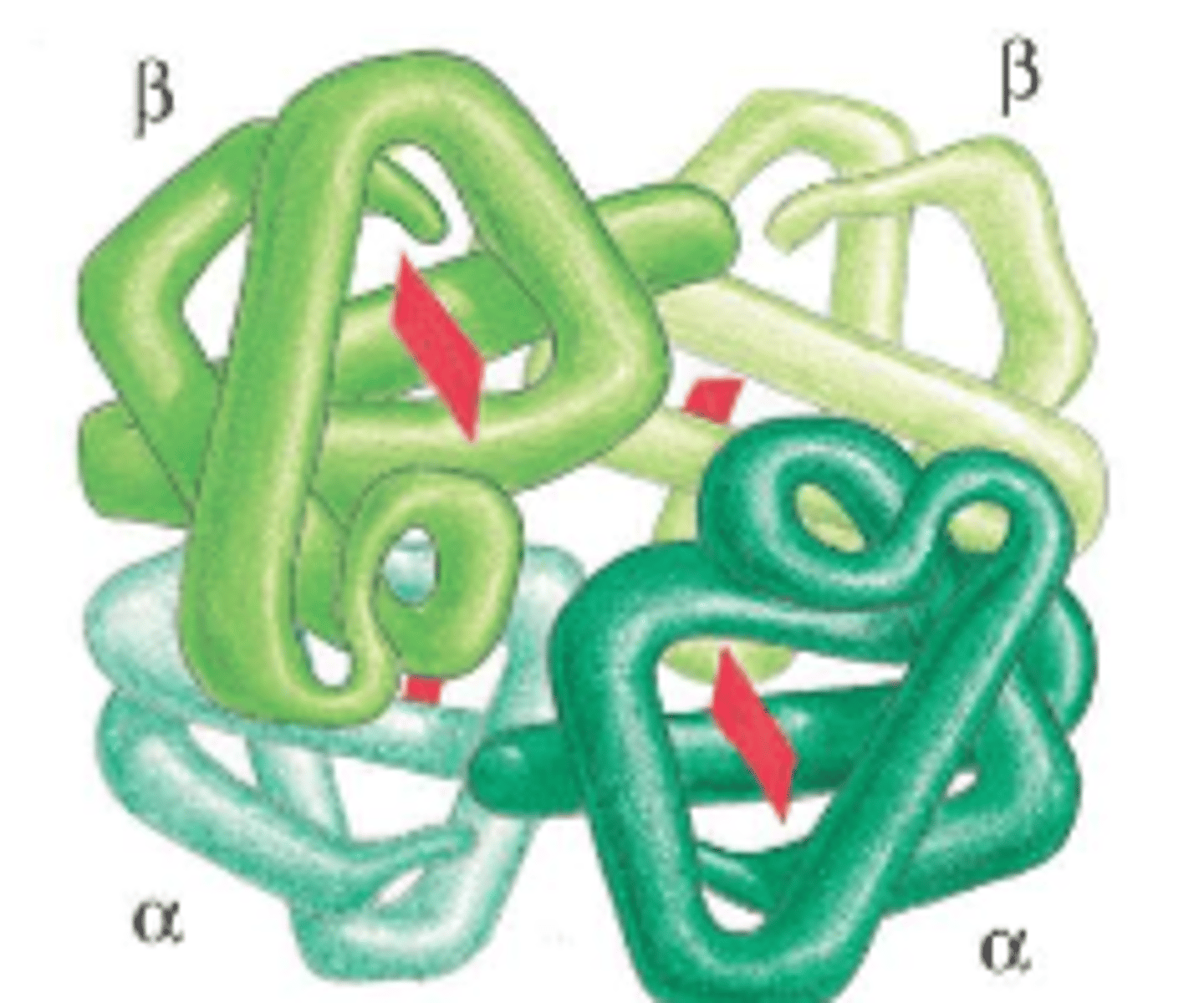
ex of protein with quaternary structure
hemoglobin
based on the structural organization, what are the two main forms of protein?
globular proteins
fibrous proteins
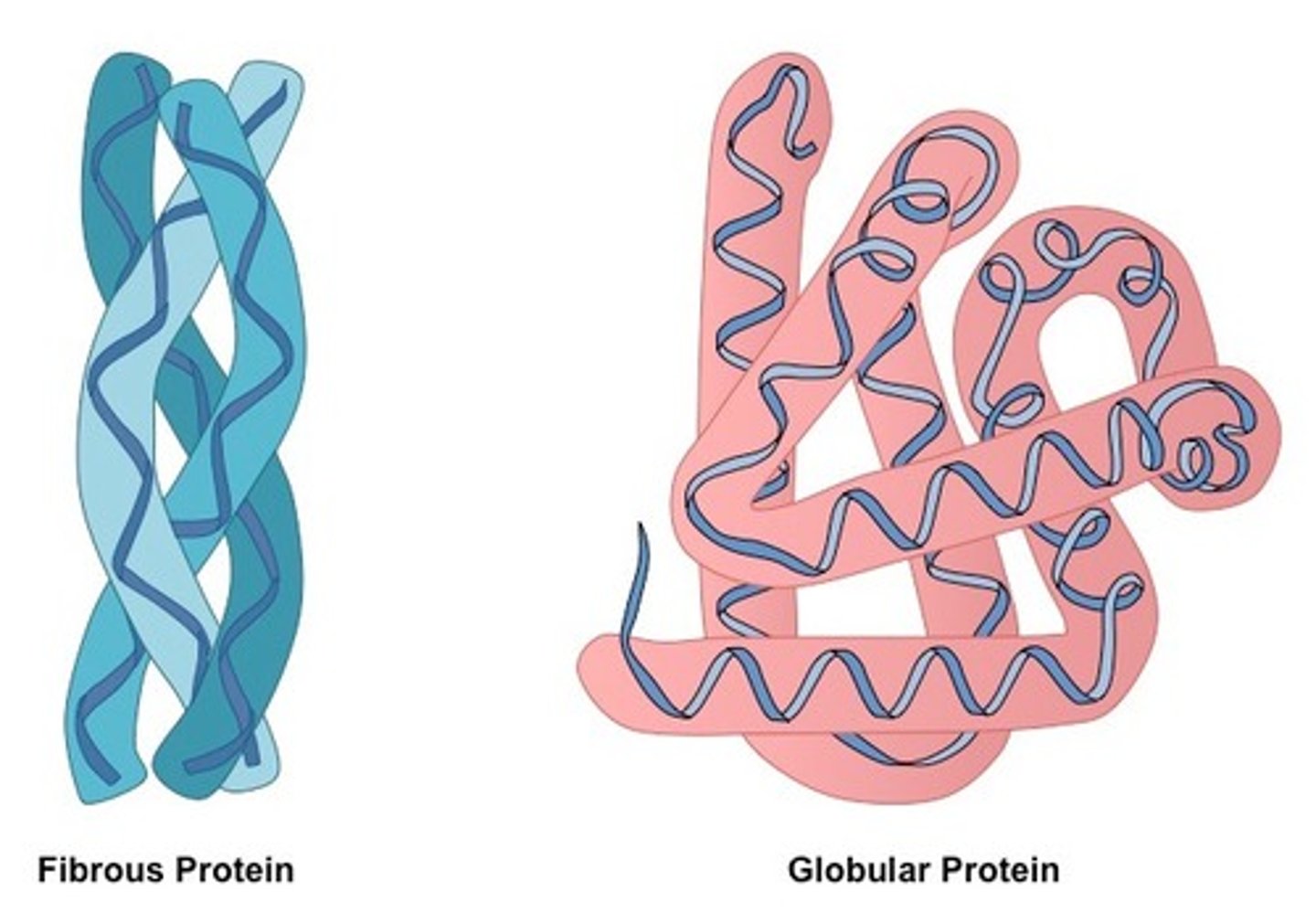
globular proteins function as
catalysts, transporters, signal transducers (functional proteins)
globular proteins, soluble or insoluble in water?
relatively soluble
role of fibrous proteins
structural rather than dynamic role

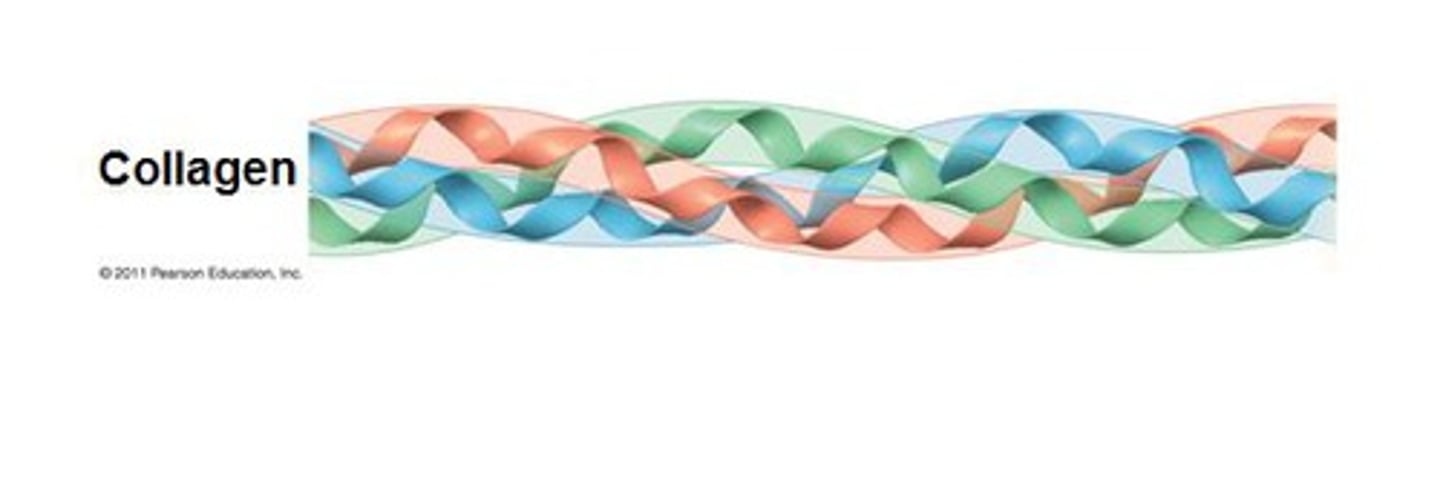
ex of fibrous proteins
collagen and keratin
fibrous proteins, solubility in water?
low solubility
properties of collagen
-provides tensile strength
-major component of connective tissues
-distinct amino acid composition: glycine, proline, 4-hydroxyproline, 3-hydroxyproline
hydroxy acids enhance
hydrogen bonding
vitamin C deficiency leads to
scurvy
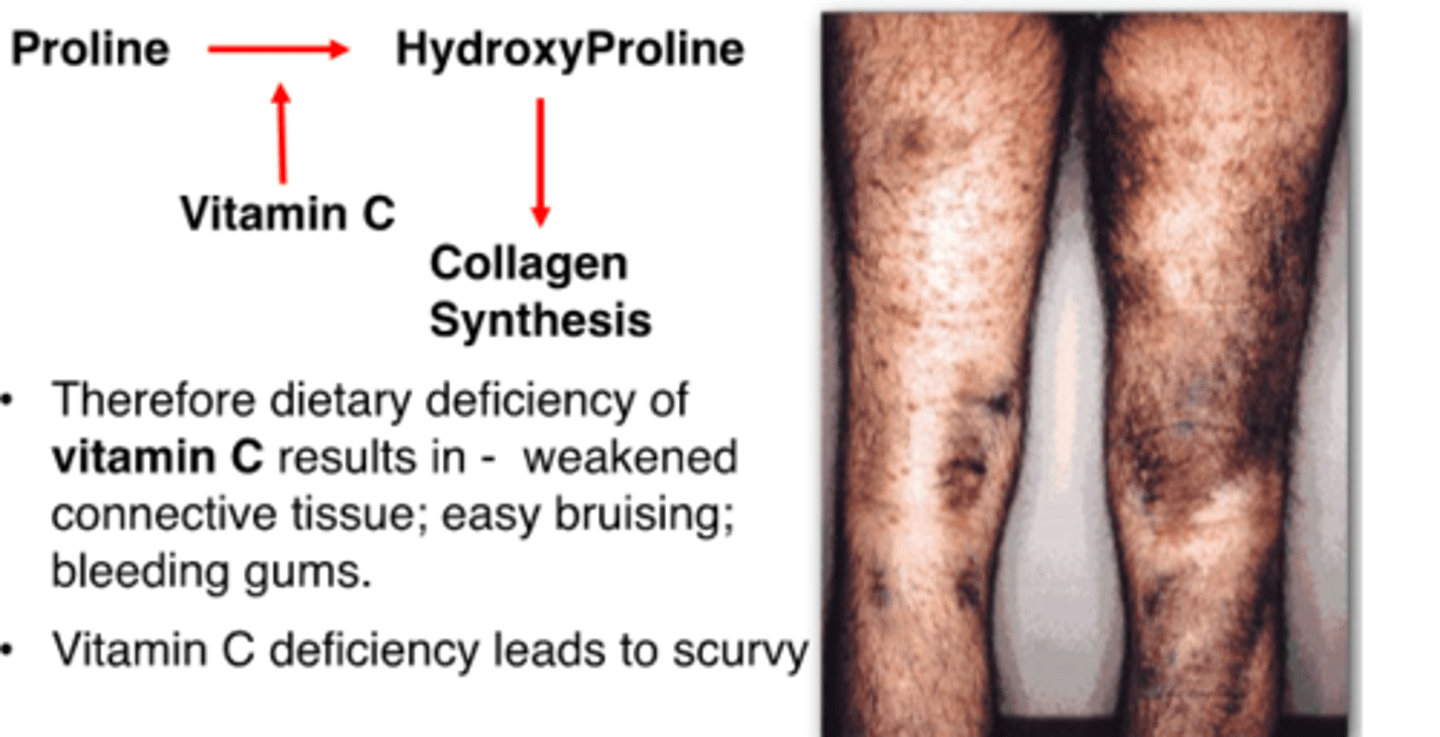
why does deficiency of vitamin C cause scurvy
-vitamin c is needed by an enzyme that converts proline to hydroxyproline
-hydroxyproline needed to form collagen
-collagen provides structural support
if Keq > 1 and delta G < 0
reaction is favorable
if Keq is < 1 and delta G is > 0
reaction is unfavorable
Keq =
[B] k1
---- = -----
[A] k-1
where B is product, A is substrate, k1 and k-1 are rate constants for forward and reverse reactions
![<p>[B] k1</p><p>---- = -----</p><p>[A] k-1</p><p>where B is product, A is substrate, k1 and k-1 are rate constants for forward and reverse reactions</p>](https://knowt-user-attachments.s3.amazonaws.com/5fd60fd0-e51b-45d1-98de-c9987f63dd2e.png)
chemical reactions have an energy barrier separating the reactants and products called the
transition state
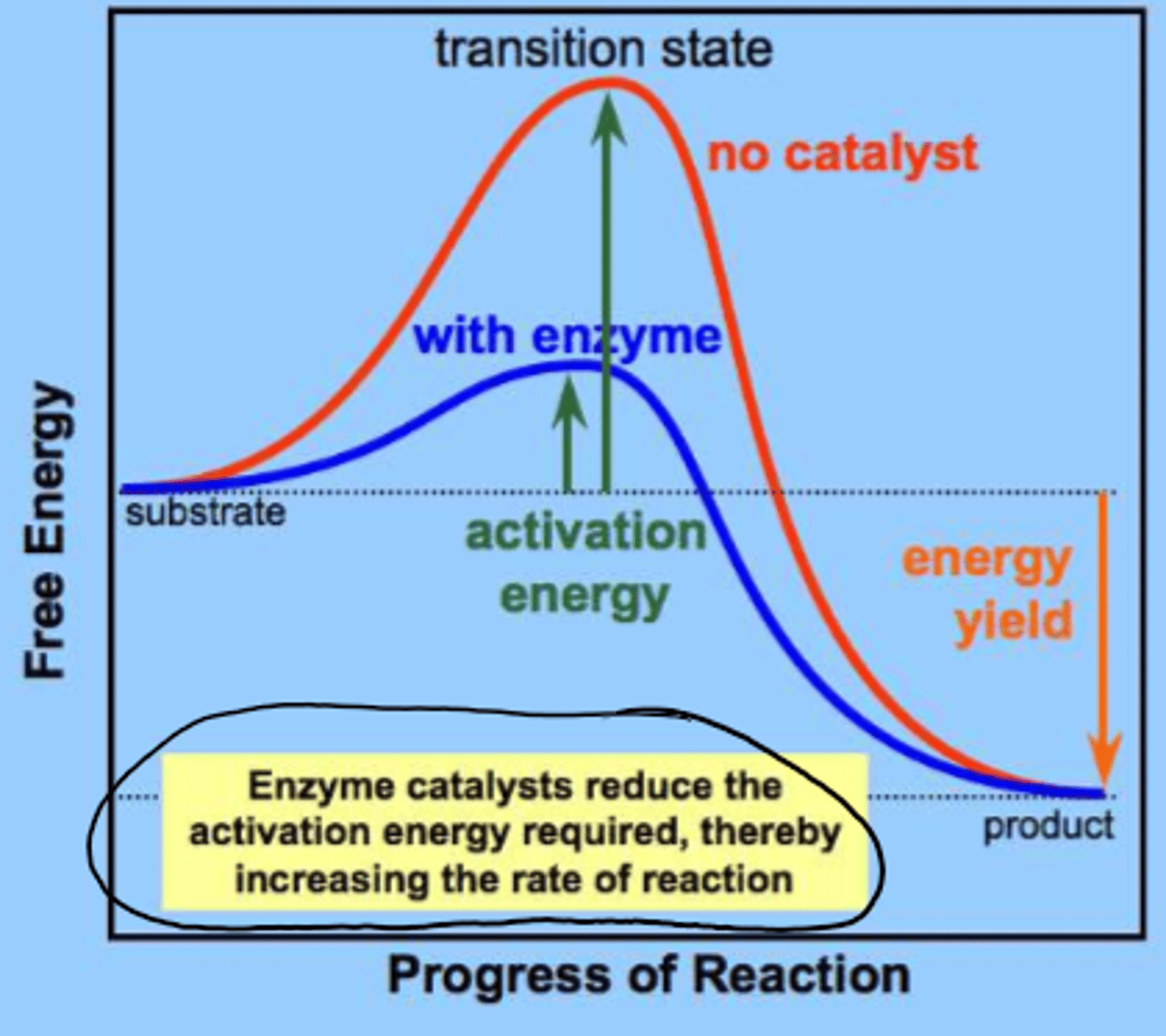
enzymes ___________ the activation energy
reduce
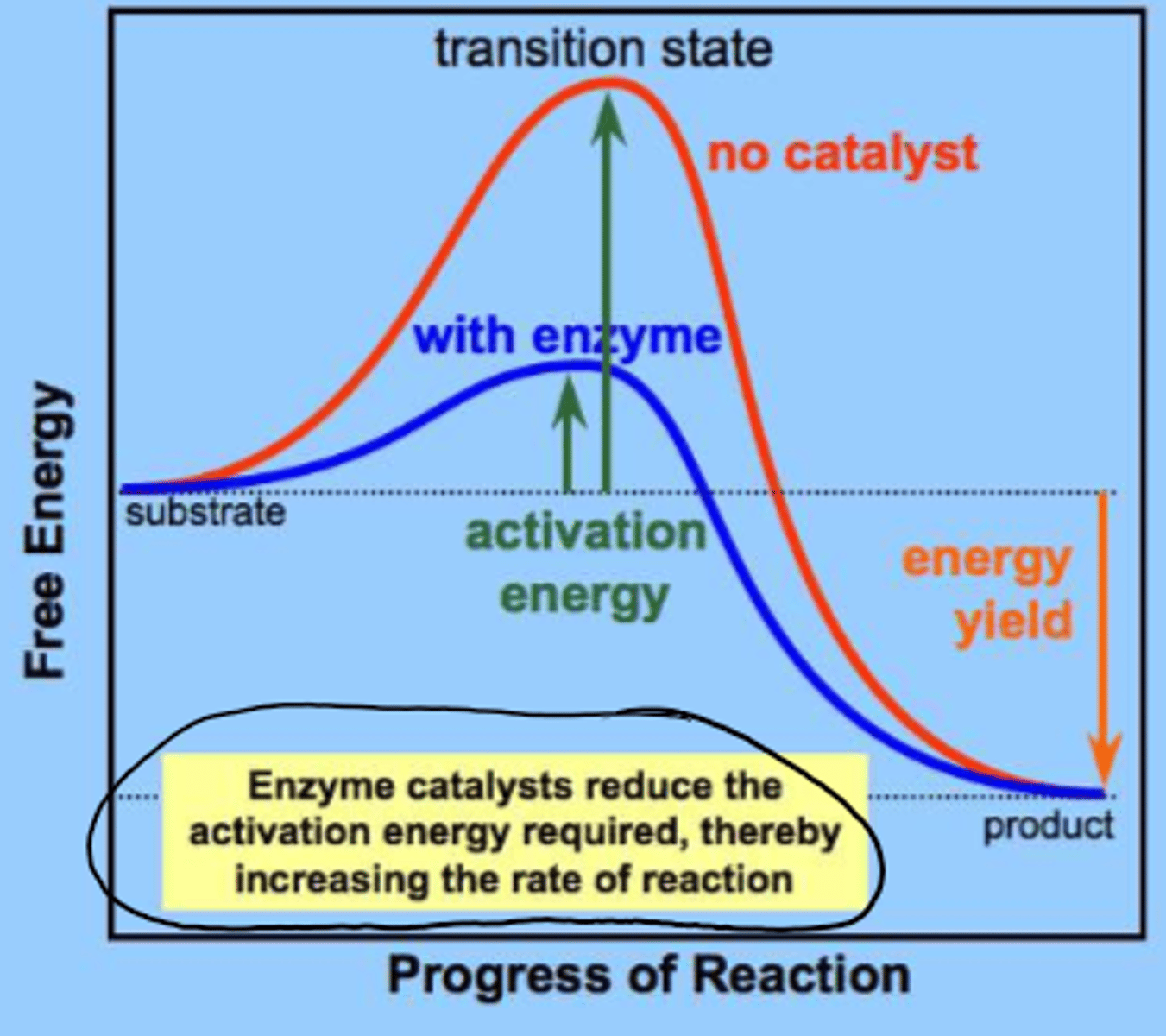
enzymes _________ rate of reaction
increase
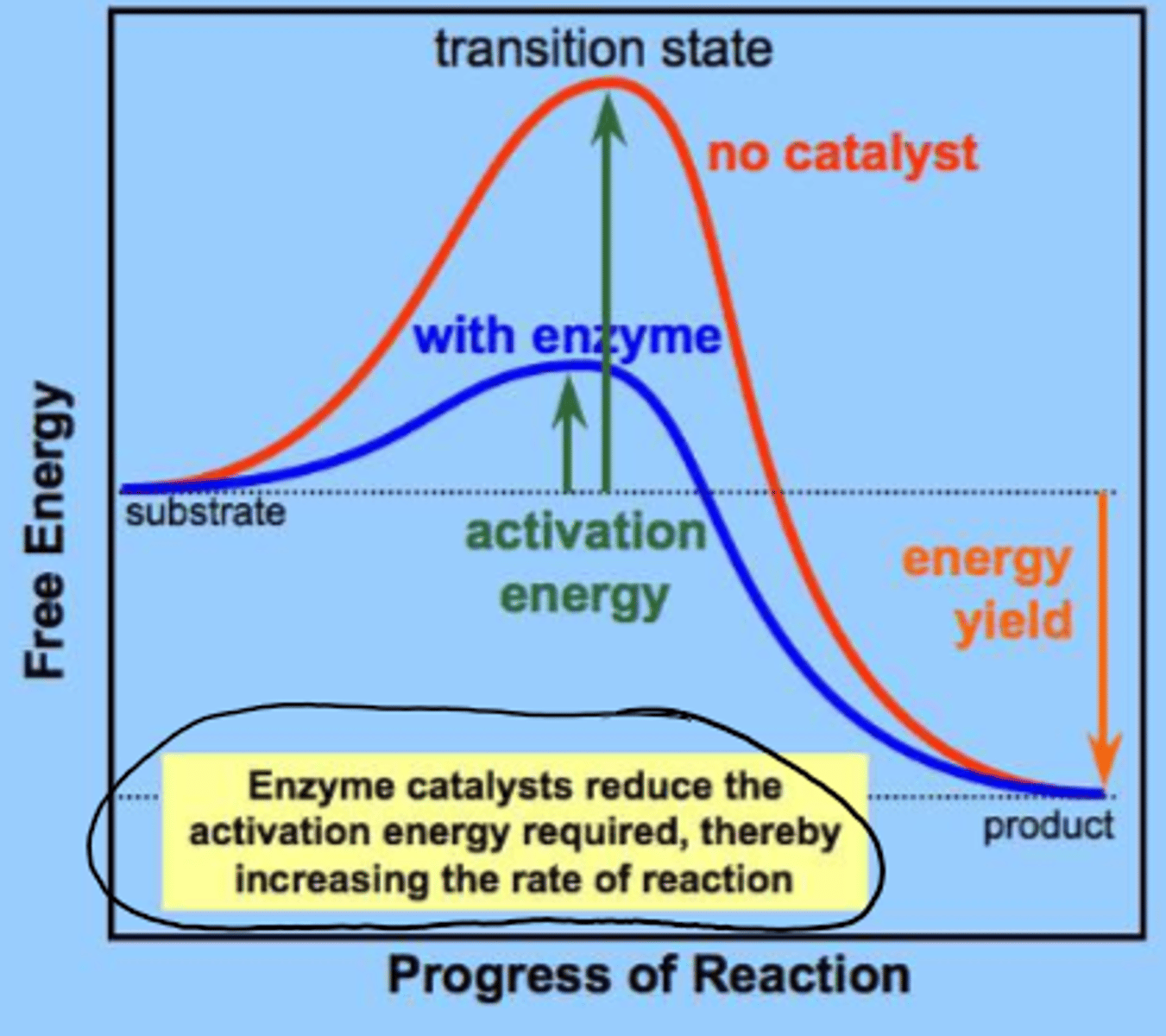
enzymes do/do not change overall delta G of a reaction
DO NOT
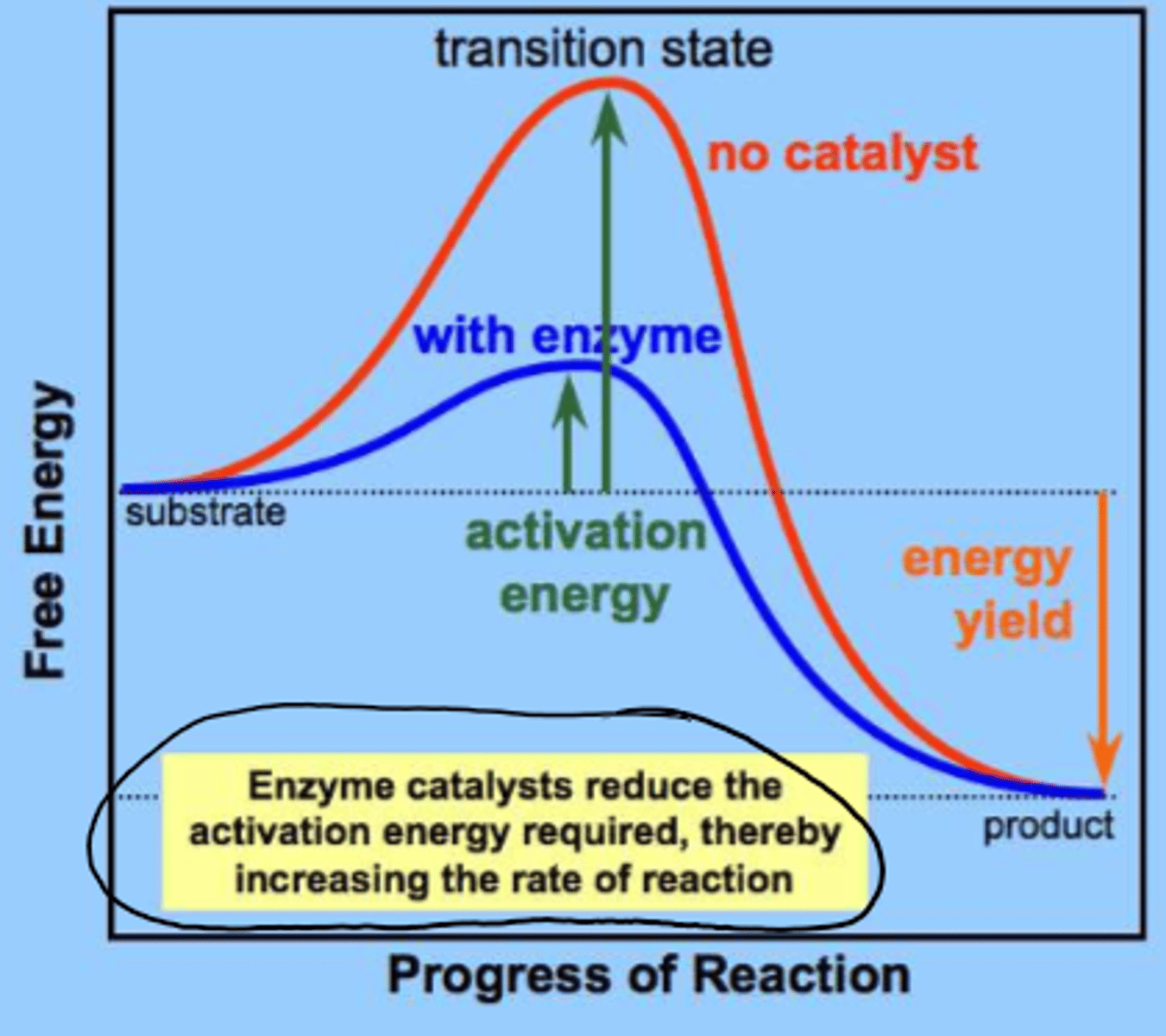
2 models of enzyme function
lock and key
induced fit model
lock and key model for substrate bonding
enzymes are able to select just ONE SUBSTRATE
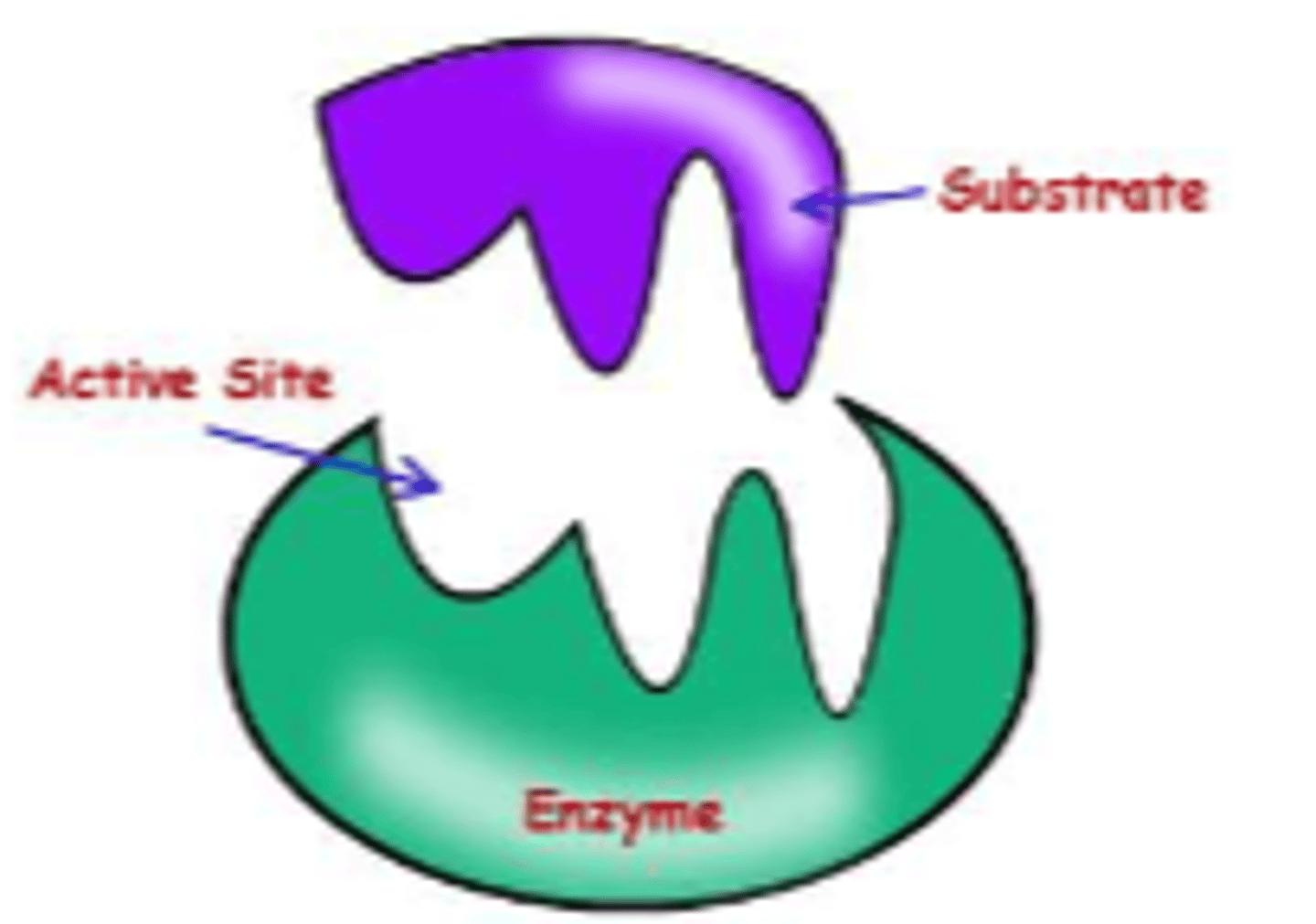
induced fit model of substrate binding
substrate binding can change the active site structure of the enzyme leading to conformational change
-flexible active site
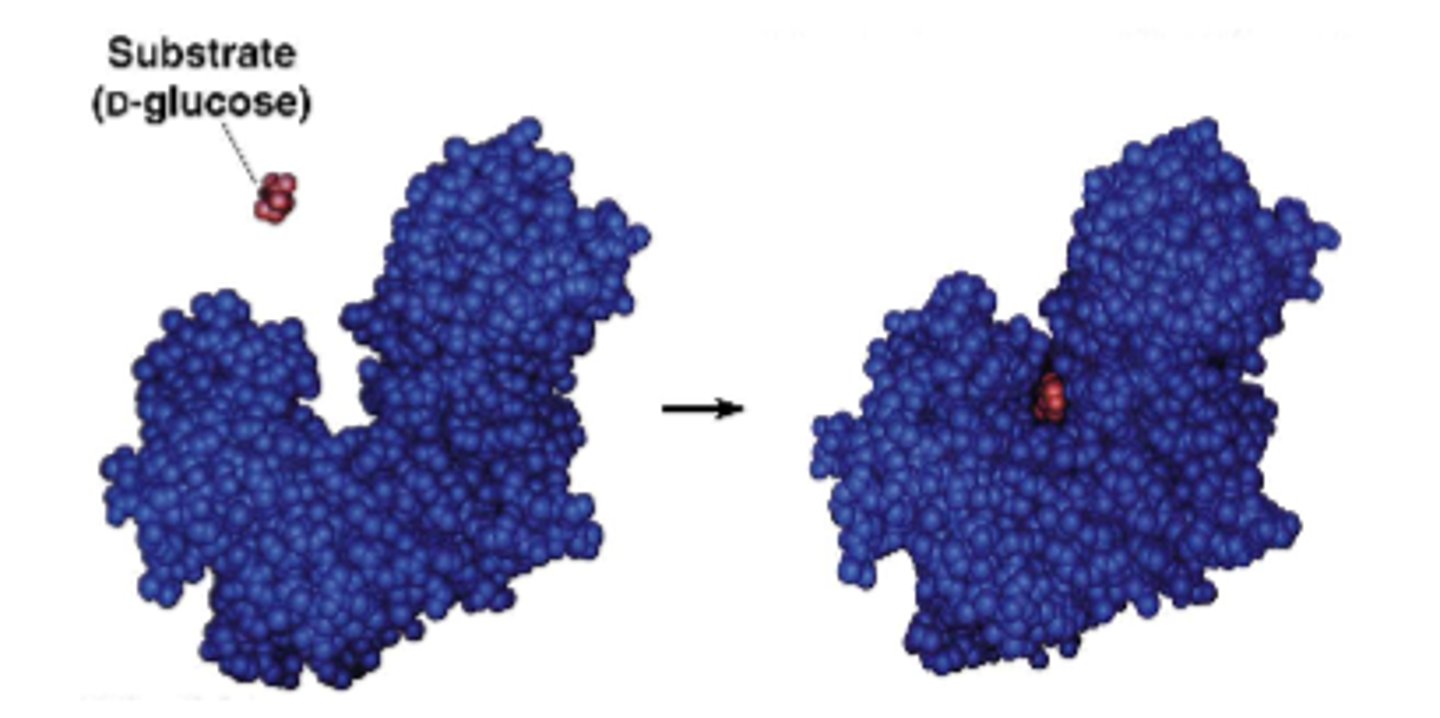
factors influencing rates of enzyme reactions
temperature
(higher=accelerates, too high will denature)
pH (can influence reaction rate, pH optimum of most enzymes)
helps in taking partially folded protein intermediates and folding them
chaperone
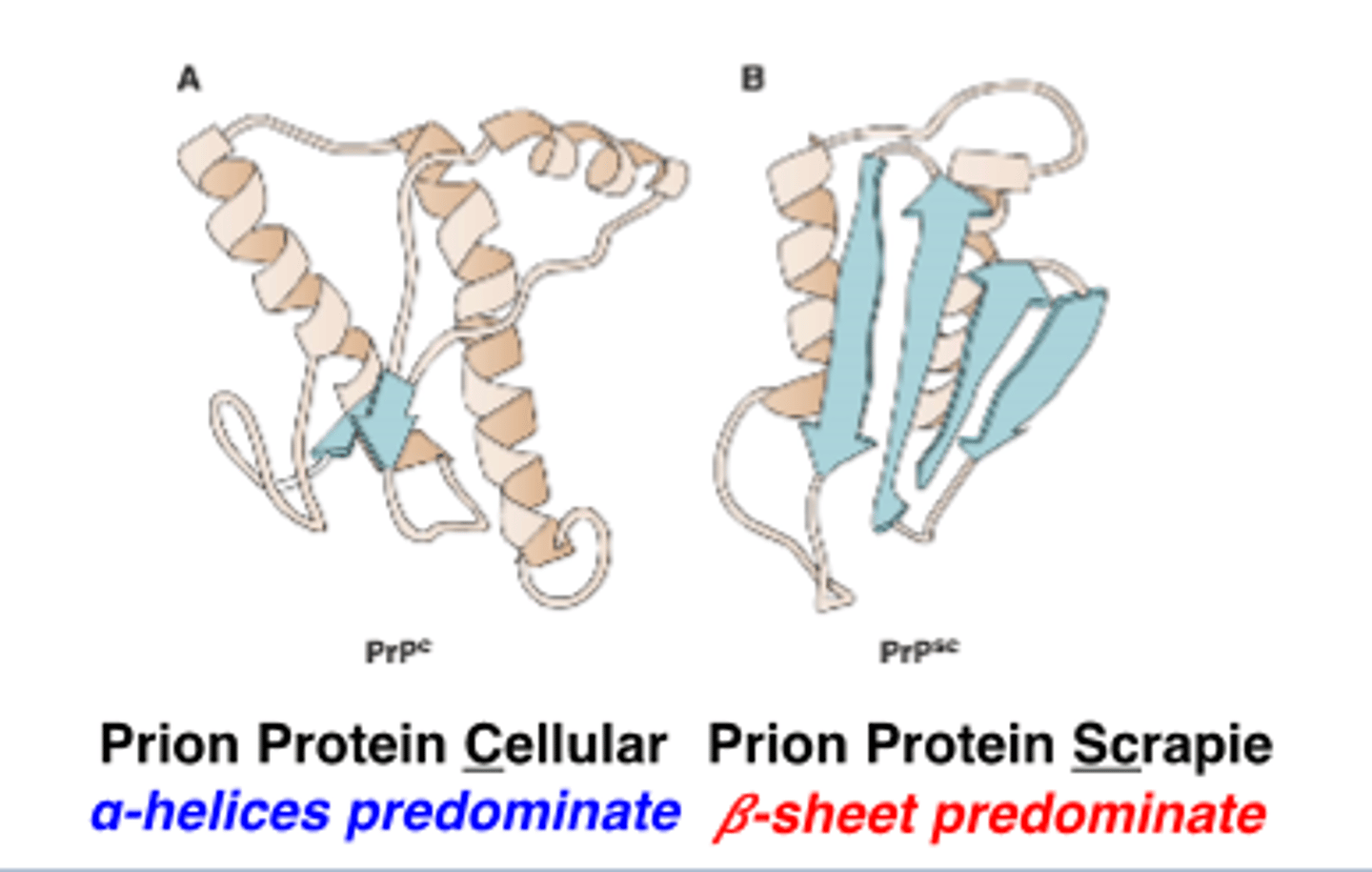
what is common amongst prion diseases
misfolded protein
prions are thought to propagate by transmitting a
misfolded protein
why do we need to study enzyme kinetics and inhibition?
drugs
drug therapies are based on ___________ enzymes
inhibiting
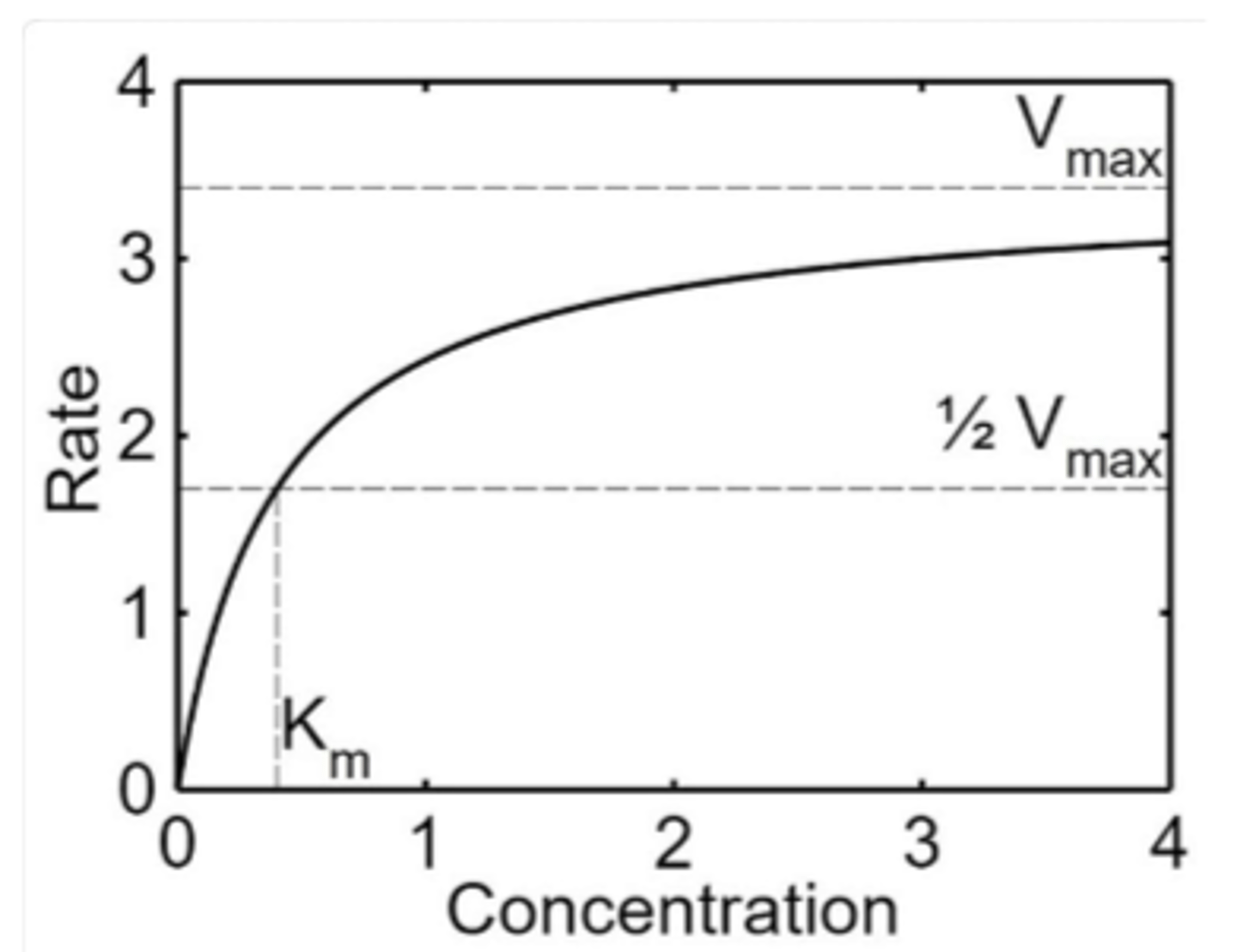
Km
Michaelis constant
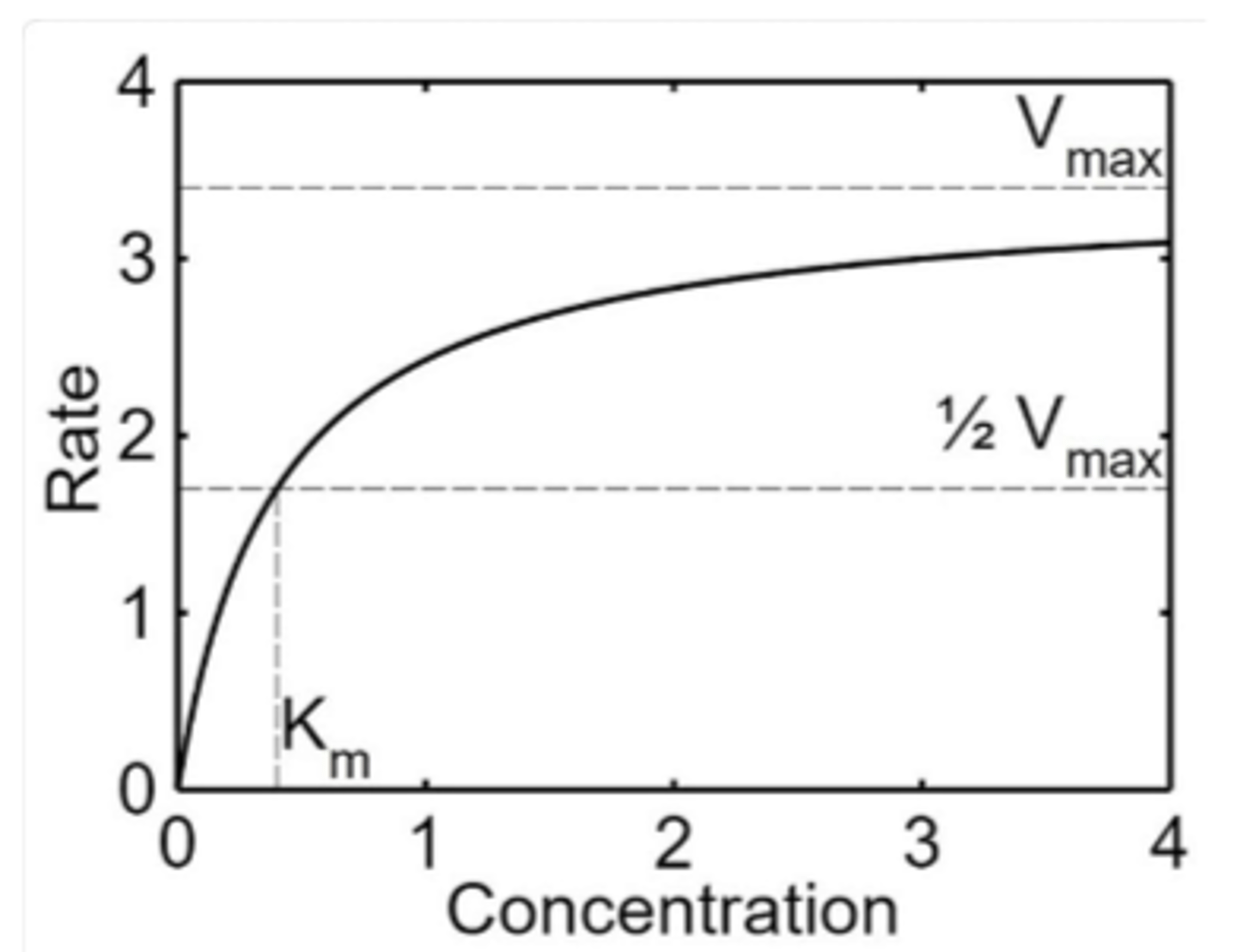
Vmax
maximum rate
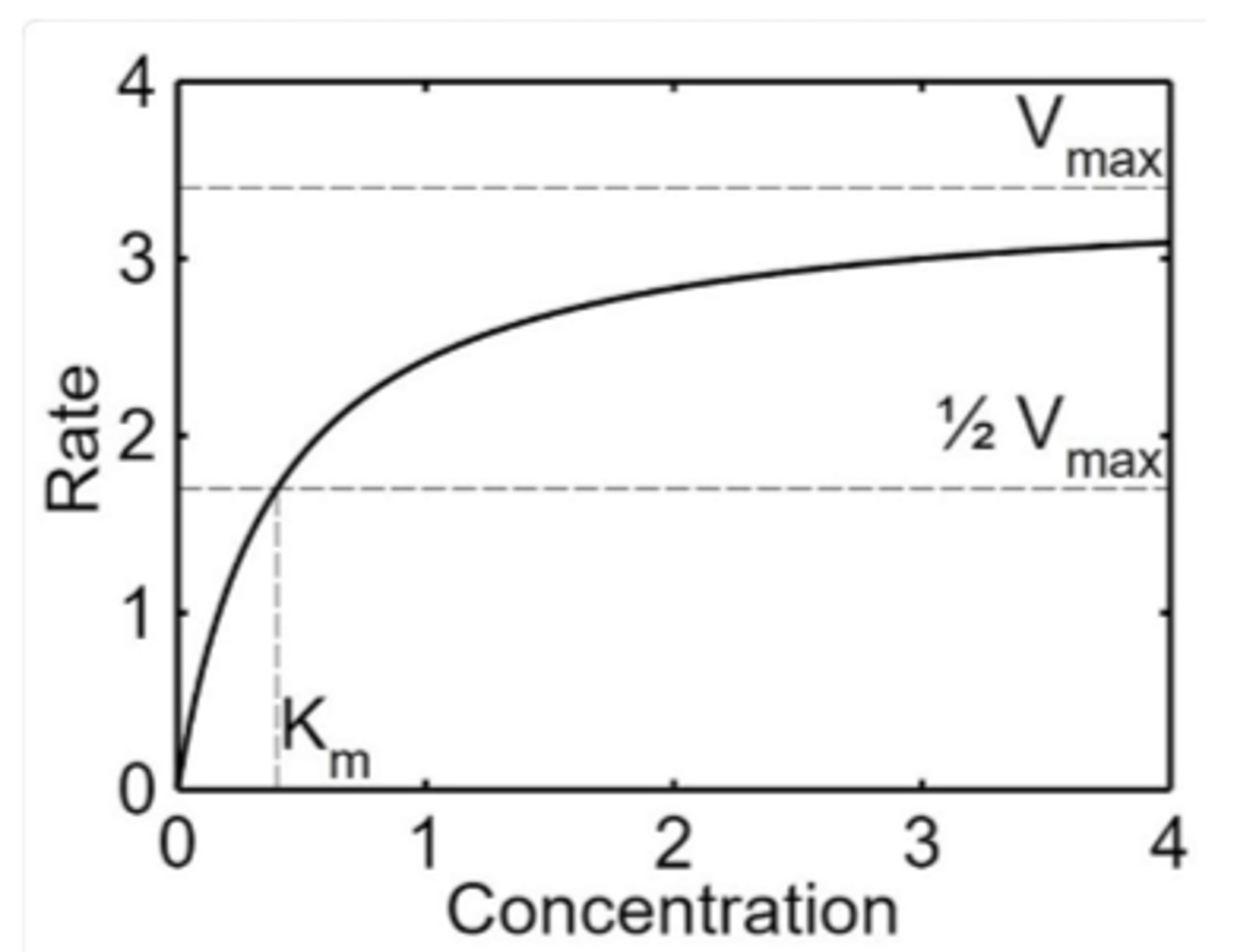
Km
affinity of enzyme for substrate
kcat
turnover number
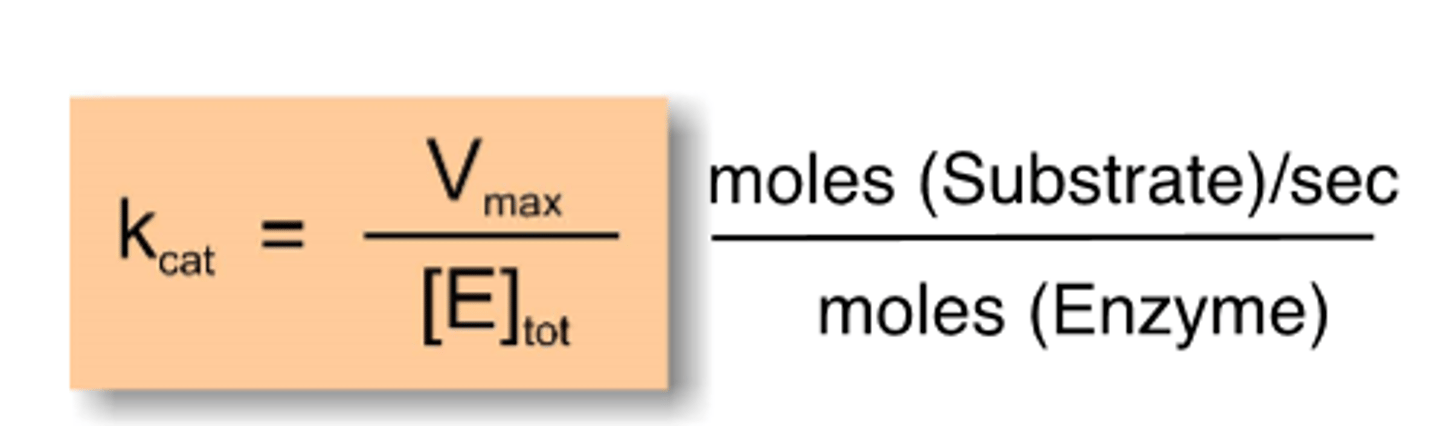
_________ Km implies a lot of substrate must be present to saturate the enzyme, meaning the enzyme has _______ affinity for the substrate
high, low
_______ Km means only a small amount of substrate is needed to saturate the enzyme, indicating a ________ affinity for substrate
low, high
enzyme inhibition types
reversible inhibitor
irreversible inhibitor
allosteric inhibitor
types of reversible inhibition
competitive
noncompetitive
uncompetitive
competitive reversible inhibitor
-inhibitor binds reversibly in the active site
-inhibitor & substrate compete for access to enzyme
-Vmax unchanged
-Km increased
-affinity decreased
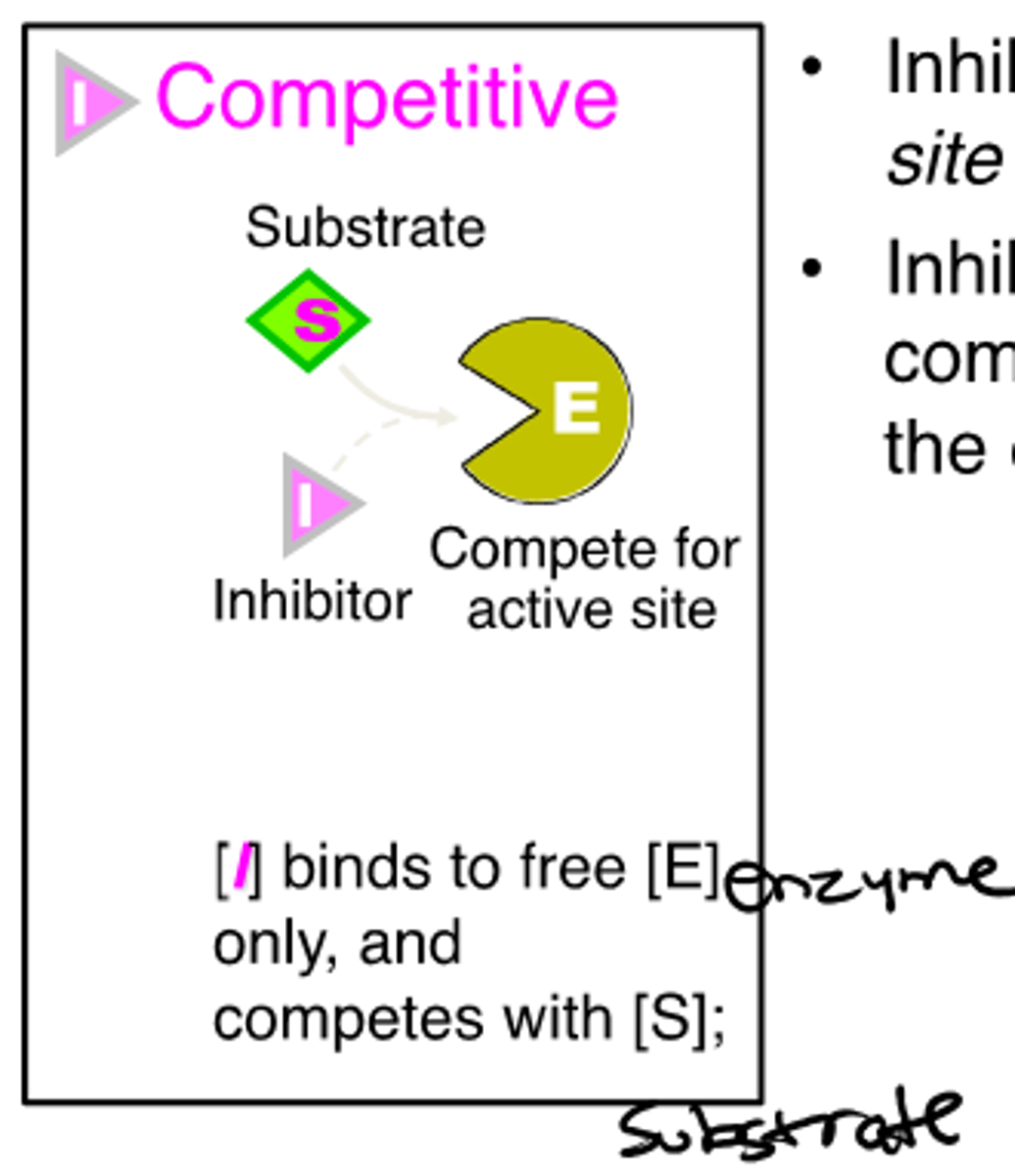
example of competitive reversible inhibitor
statins
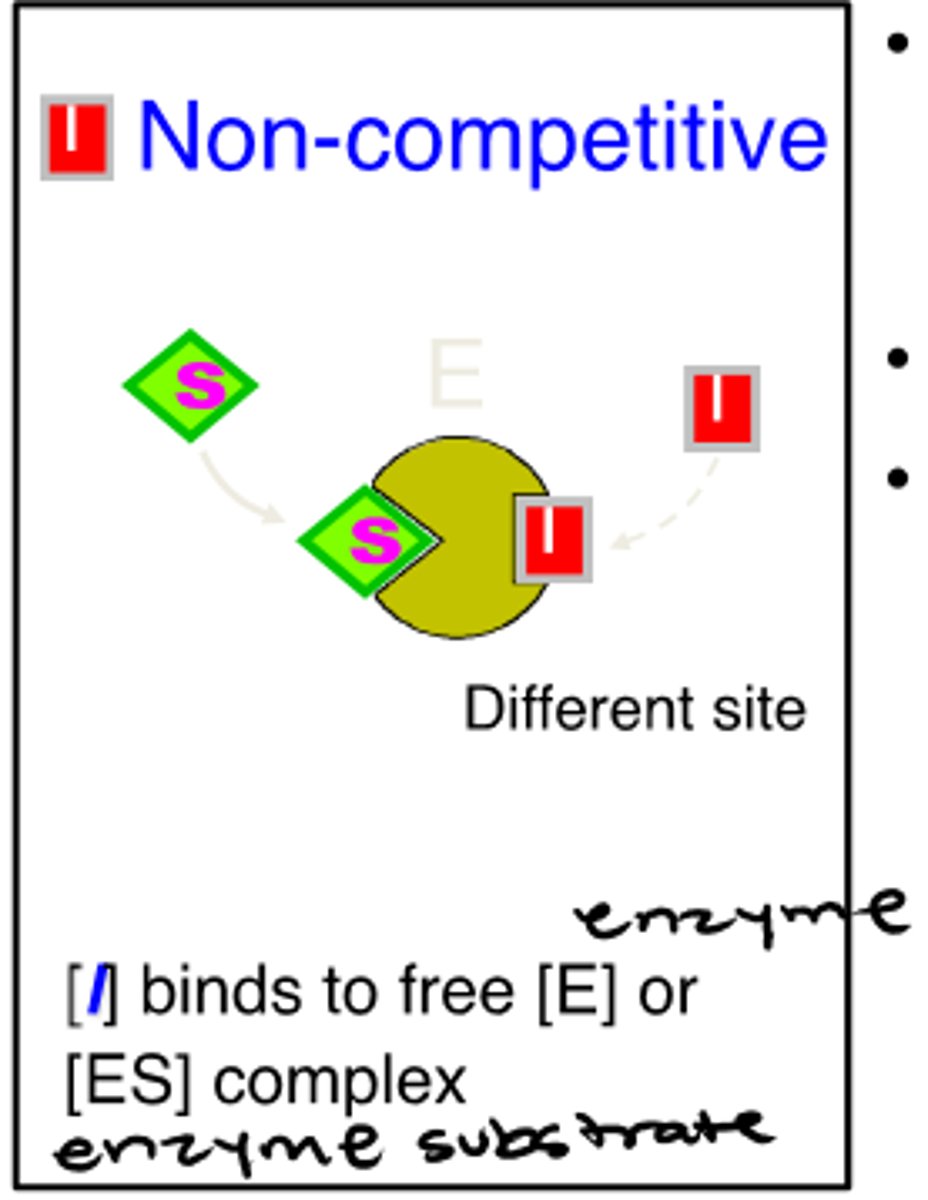
noncompetitive reversible inhibitors
-bind to a site on the enzyme distinct from the active site
-inhibitors bind to both enzyme and enzyme substrate
-Vmax decreased
-Km unchanged
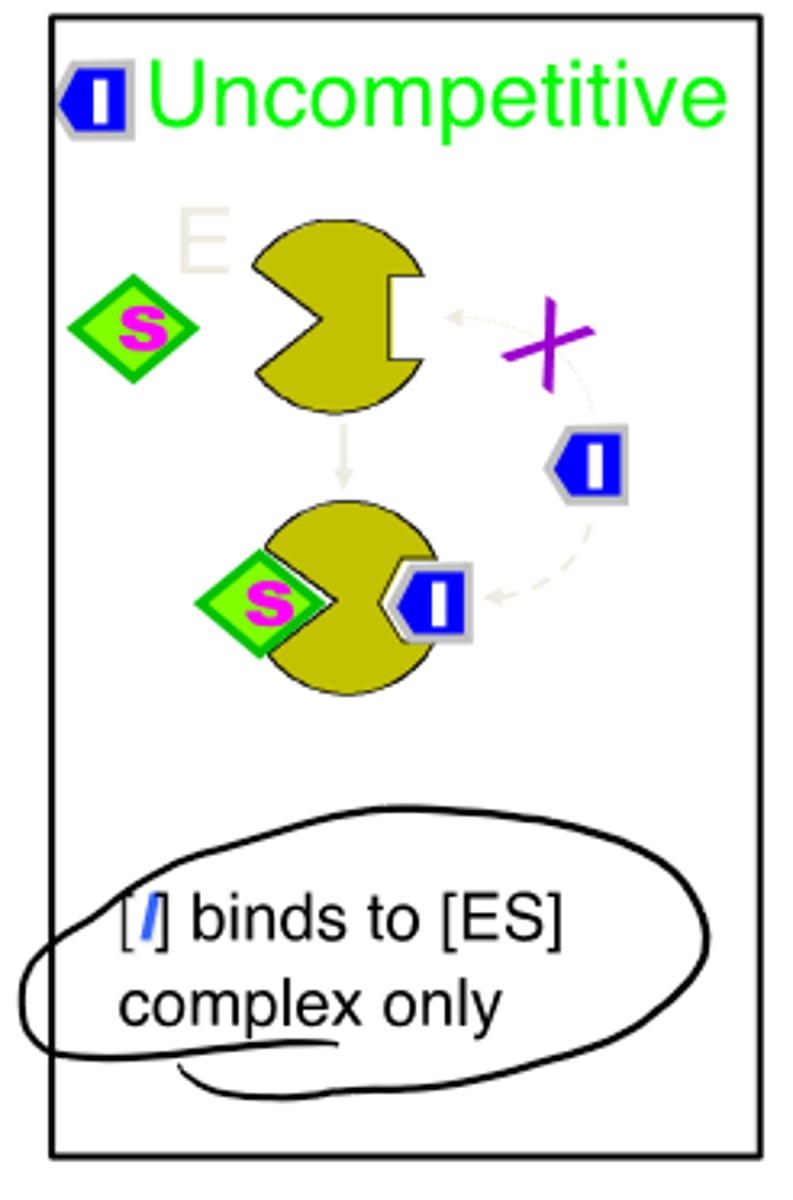
uncompetitive reversible inhibitors
-binds to enzyme-substrate complex only
-both Vmax and Km decreased
irreversible inhibitors
-suicide inhibitors
-covalently modify the enzyme, destroying its activity permanently
example of irreversible inhibitor
aspirin
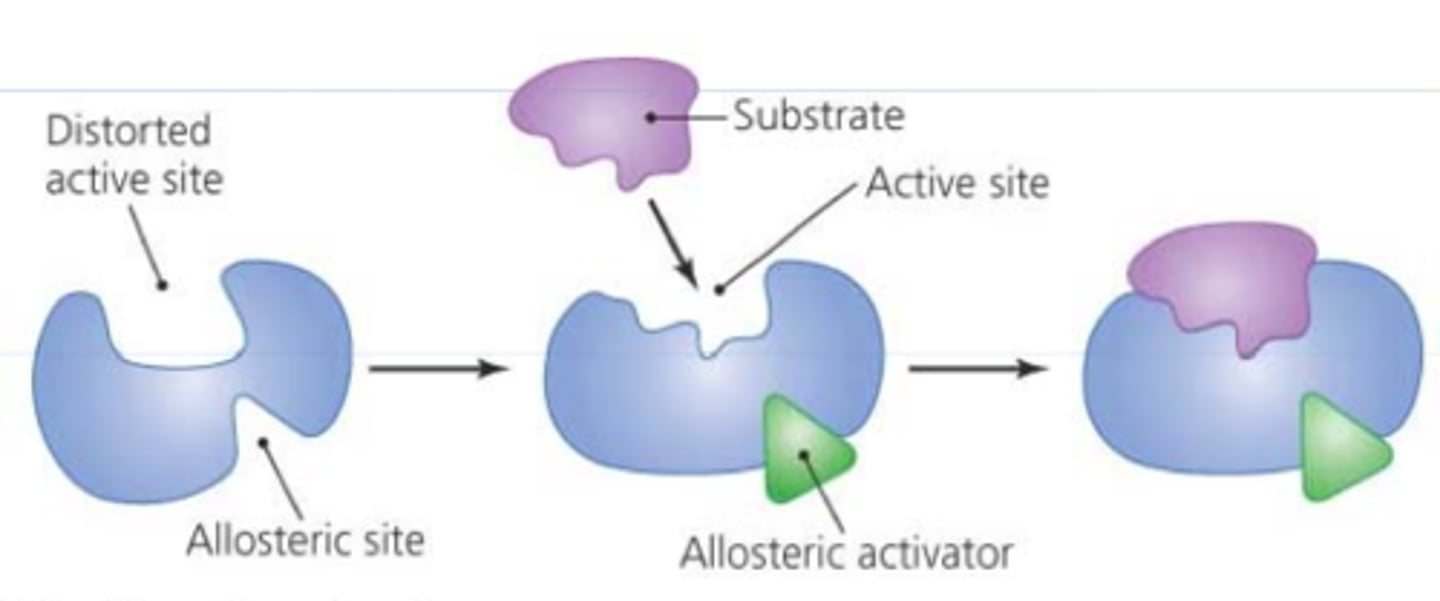
allosteric inhibitor
-remote control
-inhibitor binds to different site than active site and control the active site (inhibitor binds and changes active site- less active)
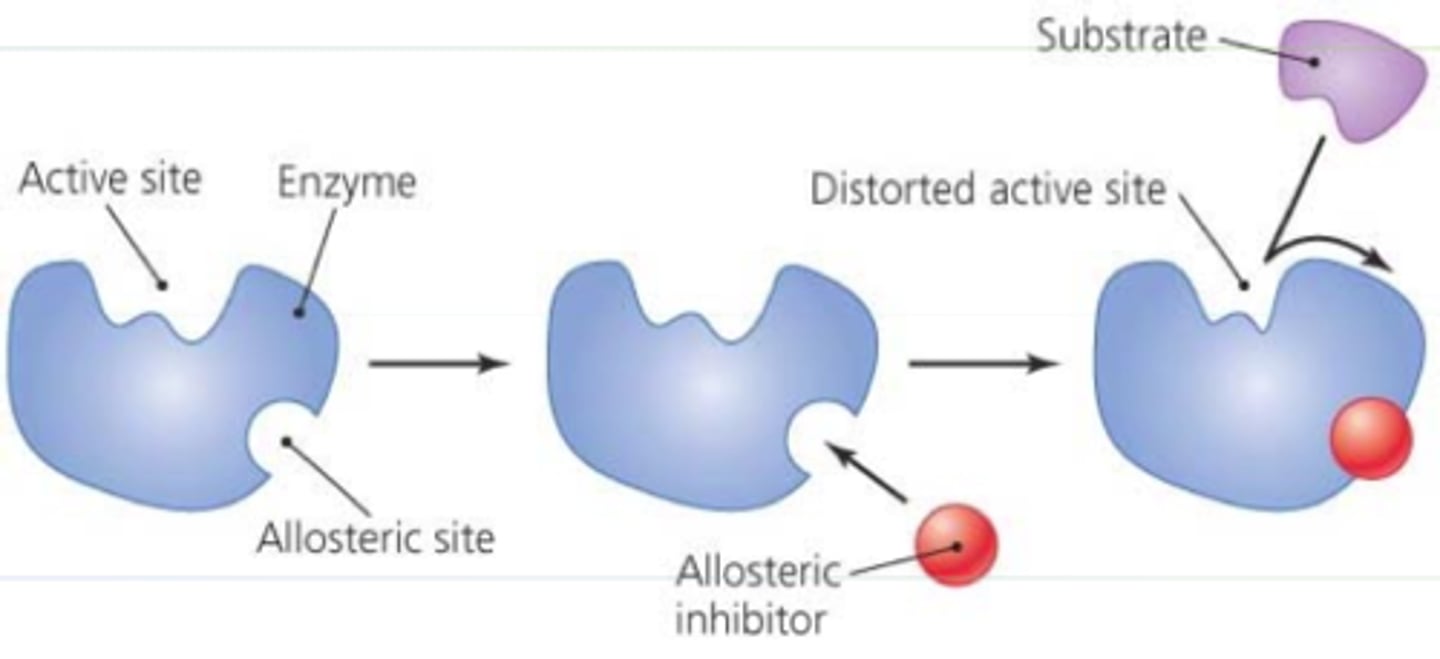
allosteric activation
-makes it faster
-produces more products
allosteric inhibition show ___________ curve
sigmoid
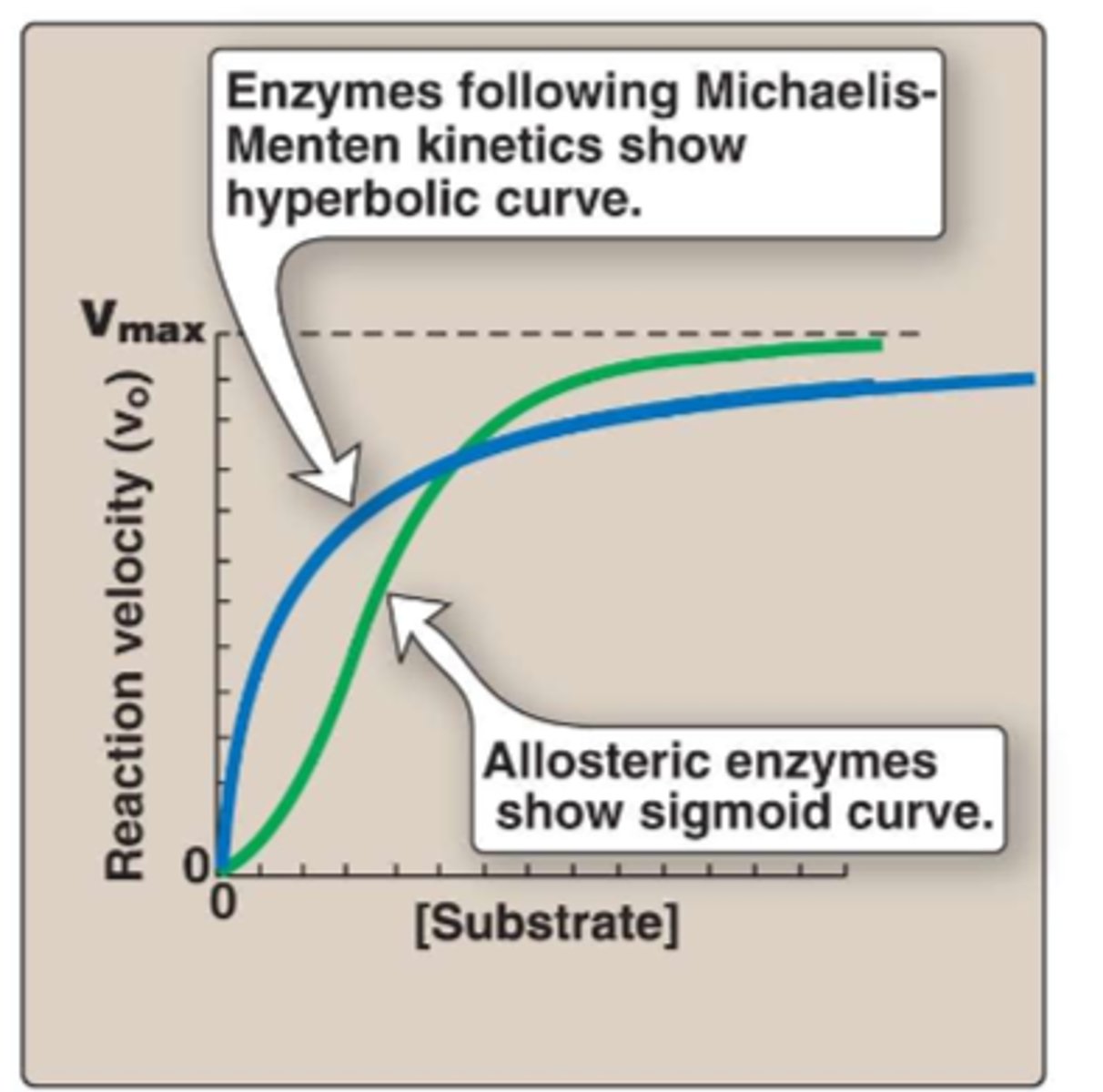
maturity onset diabetes of the young (MODY) can be caused by mutations that either
1. increase the Km (poor affinity for ligand)
2. decrease the Vmax (poor rate of conversion)
3. affect both Km and Vmax of glucokinase
-therefore the mutation of the enzymes cause their Km and Vmax to change, affecting glucose transport
3 stages of signal transduction
1. Reception
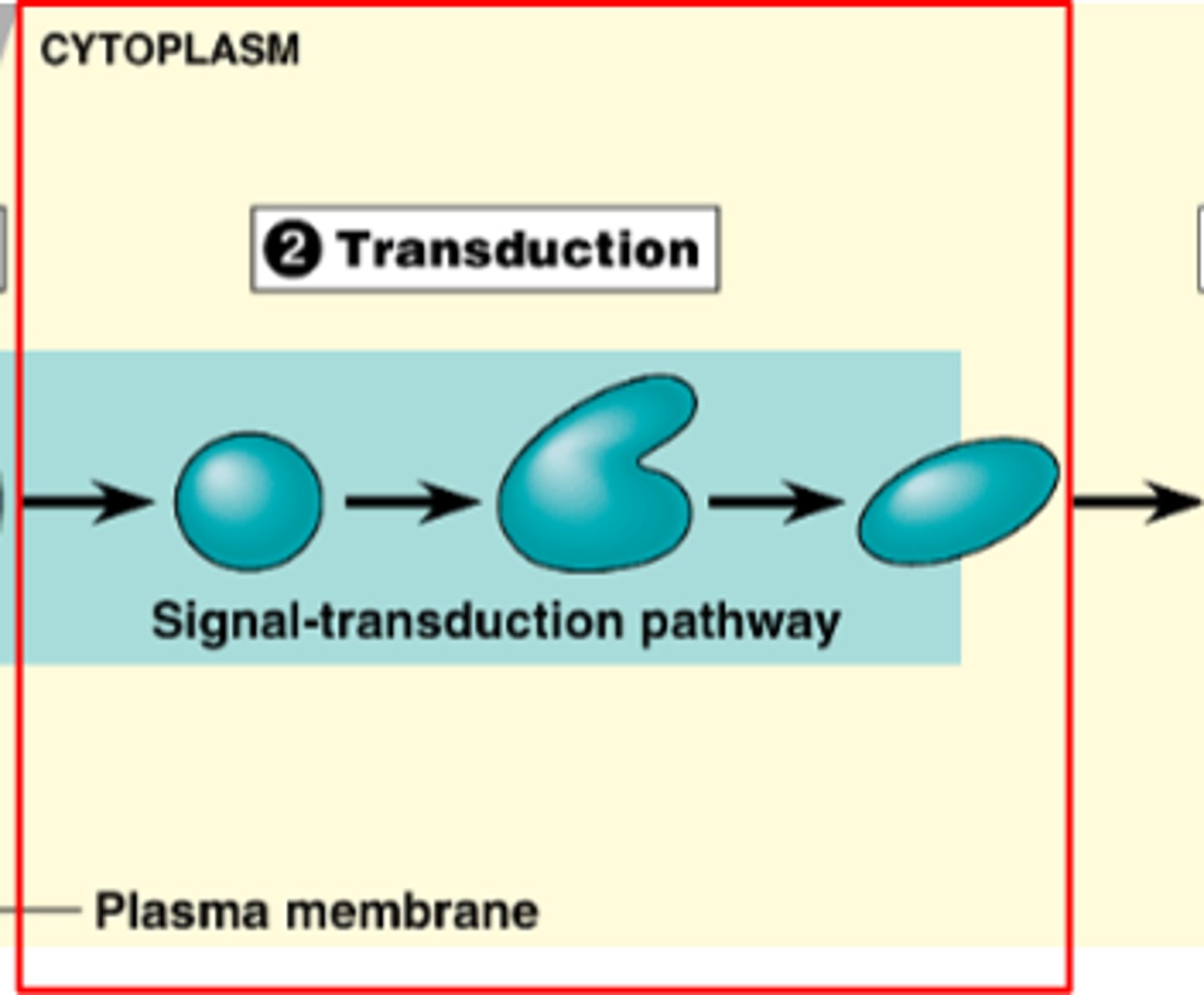
2. Transduction
3. Response
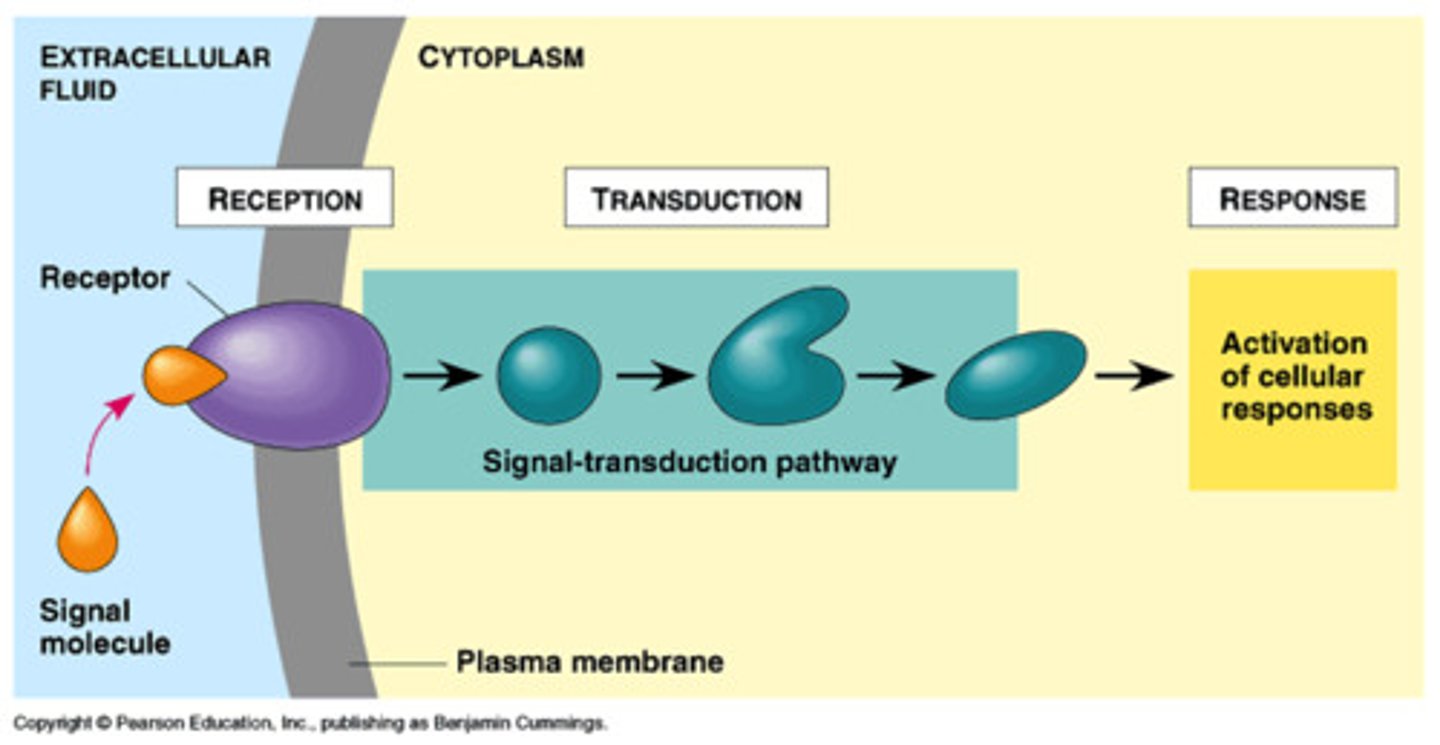
reception of extracellular signal by cell
needs receptor, binds to ligand
transduction
transduction of signal from outside of cell to inside of cell, often multi-stepped
cellular response
occurs entirely in receiving cell
cells that produce the signaling molecule referred to as
signaling cells
cells that receive the signal are
target cells
4 types of intercellular signaling
1. Contact-dependent
2. Paracrine
3. Synaptic
4. Endocrine
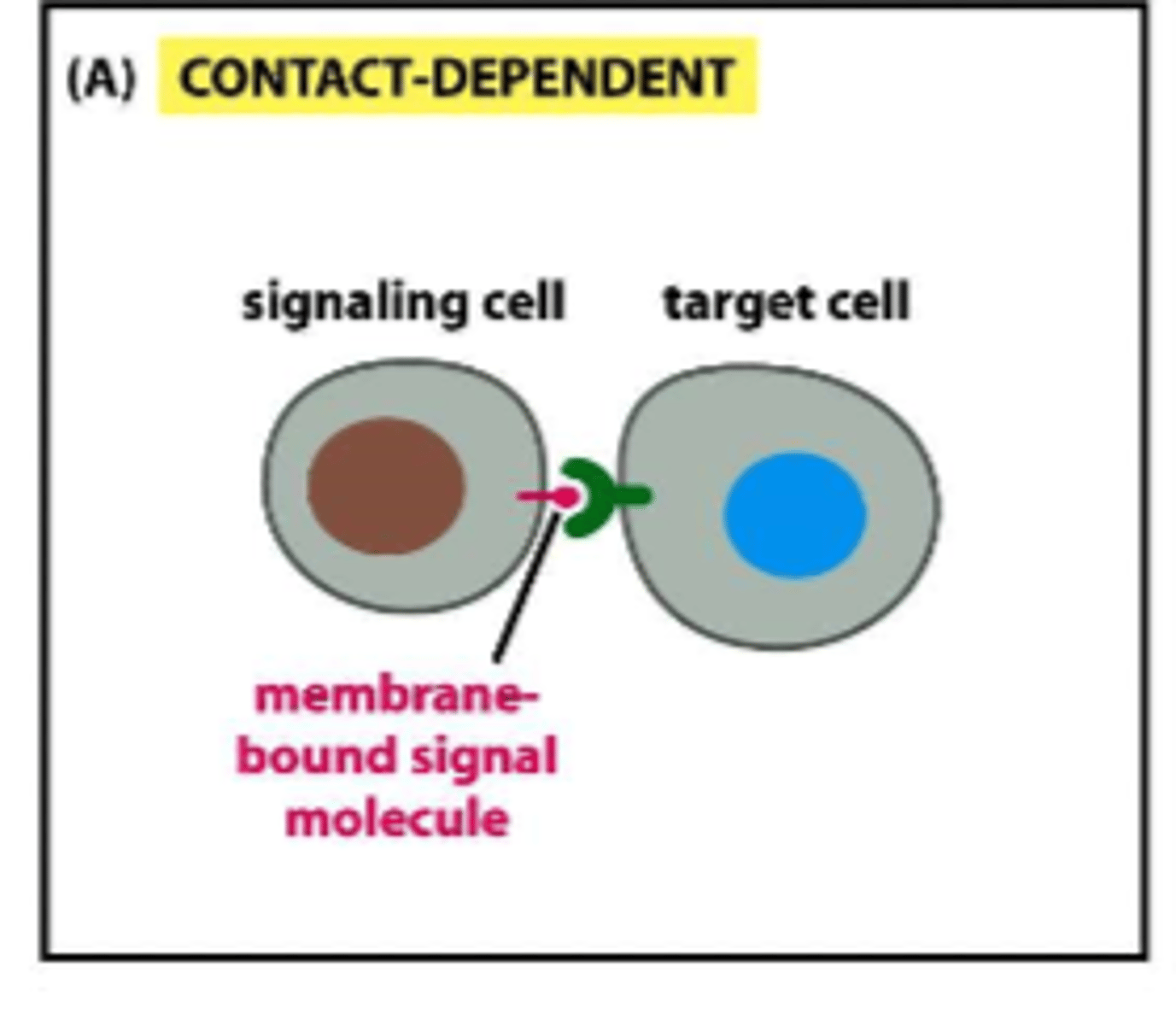
contact-dependent intercellullar signaling
very close to each other
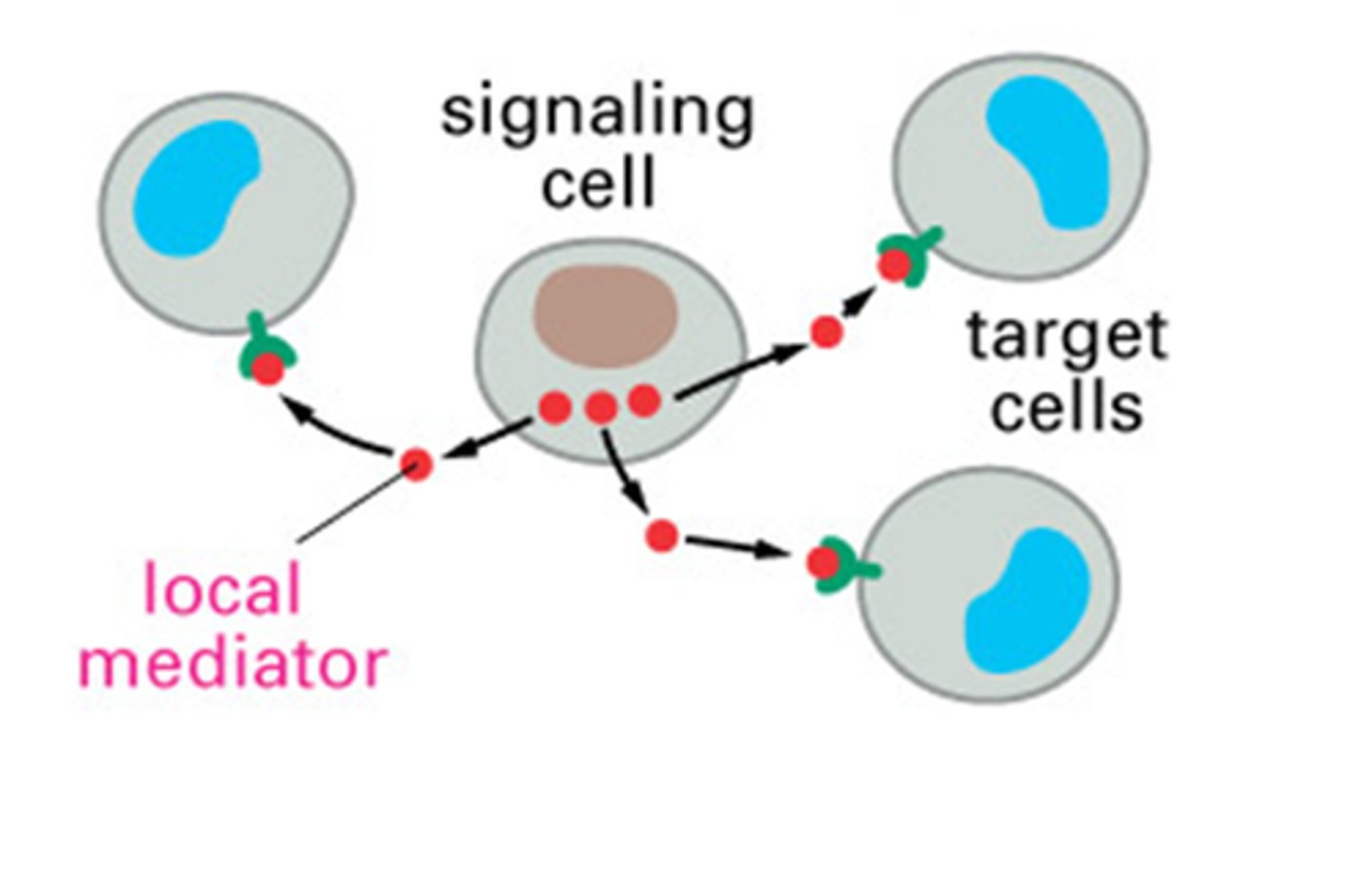
paracrine intercellular signaling
one cell makes molecules
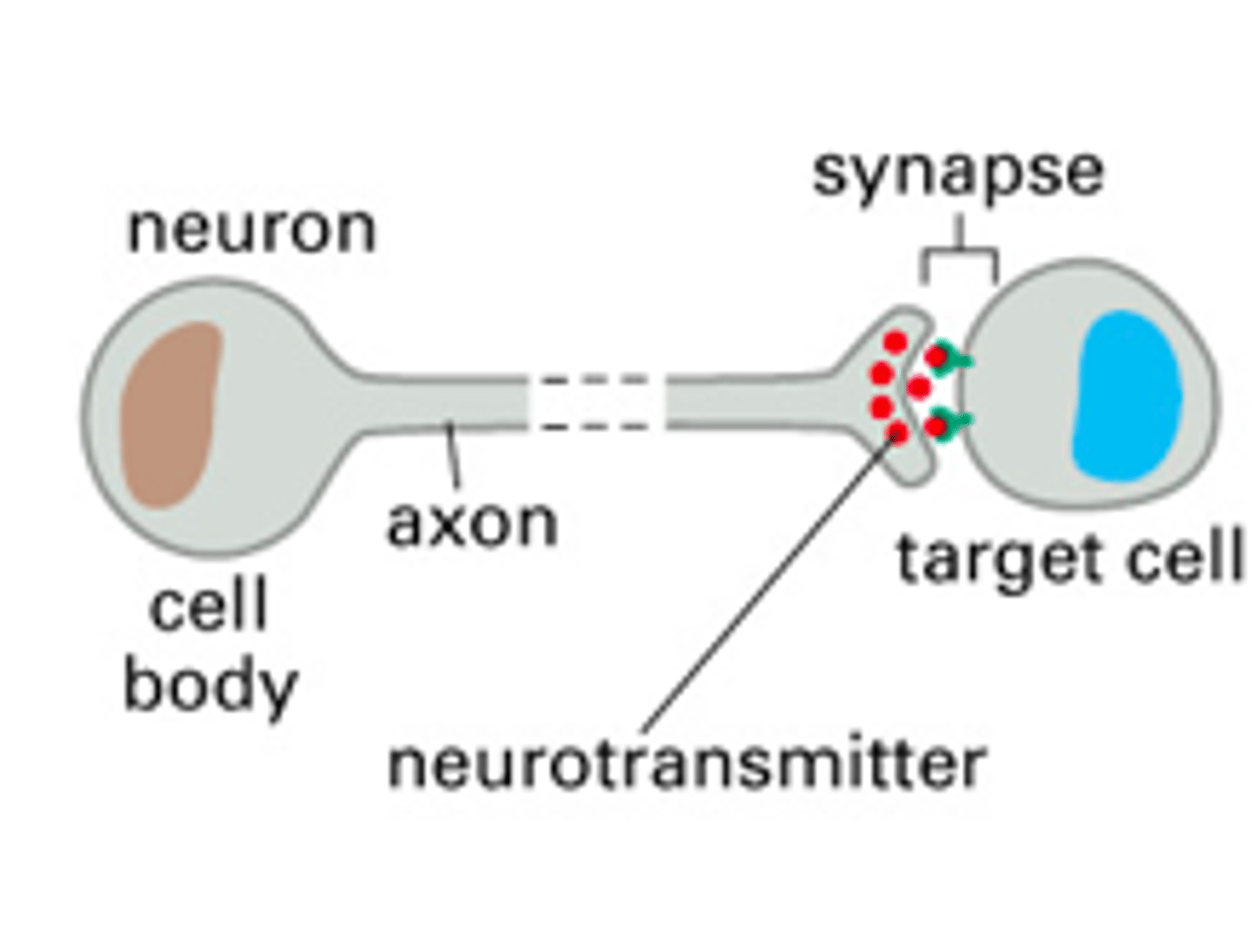
synaptic intercellular signaling
synapses released
endocrine intercellular signaling
stable molecules released into bloodstream via endocrine cells
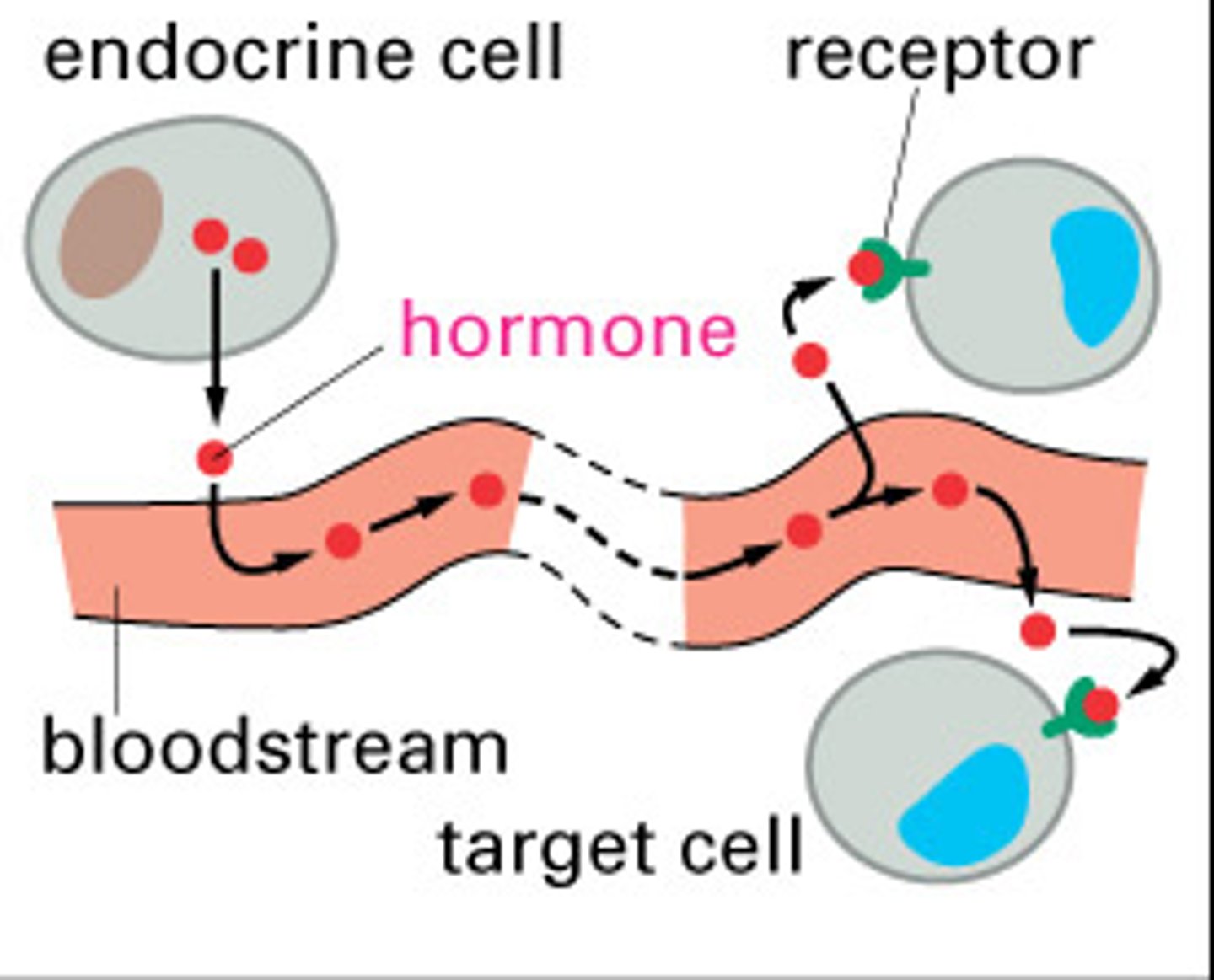
-largest class of signaling proteins involved in many biological process and pathologies
- about 50% of all modern drugs target them
G-protein coupled receptors (GPCRs)
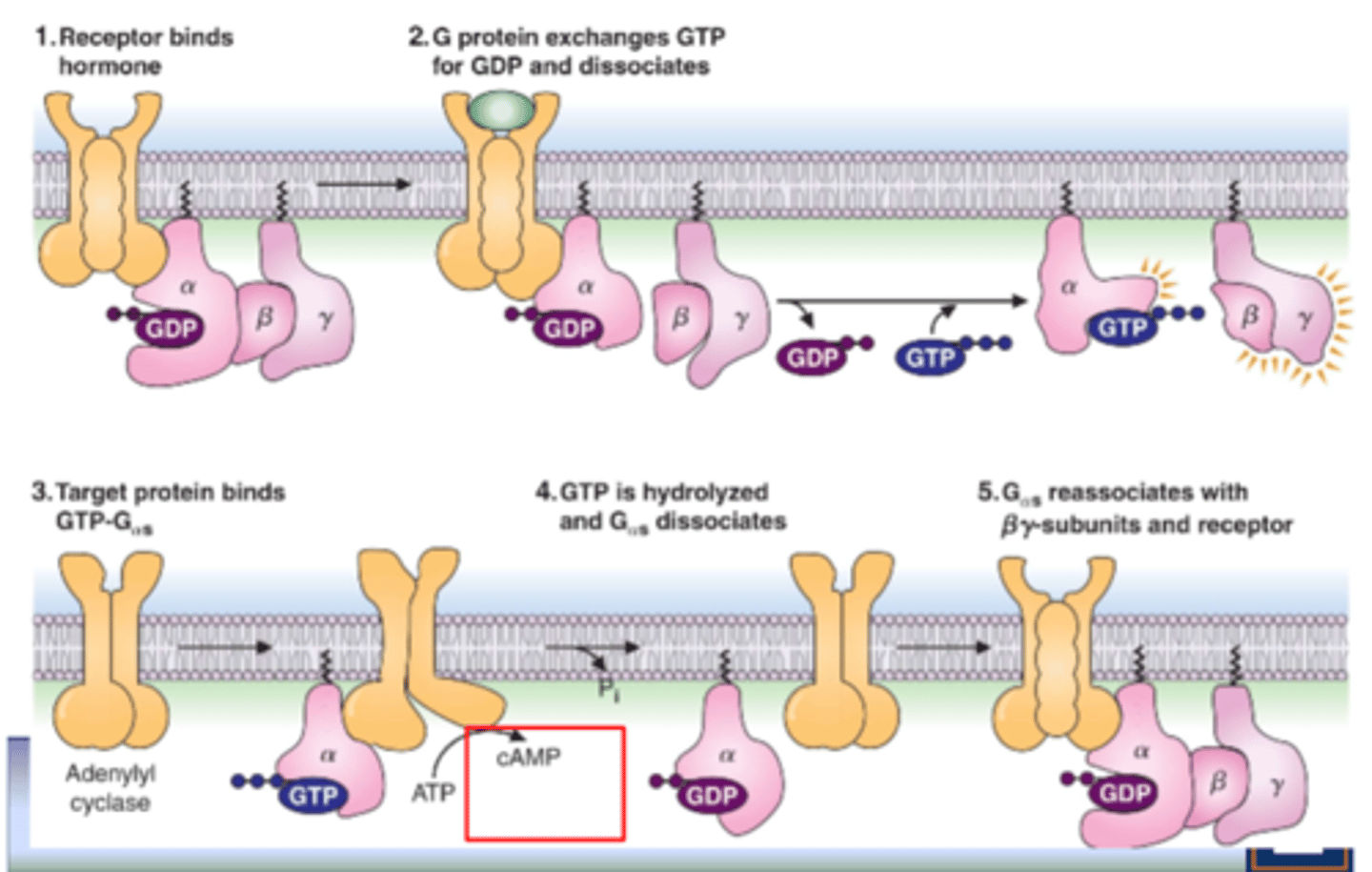
signal transduction by GPCRs
1. receptor binds to G proteins (a, B, Y) that contain GDP bound to a
2. G protein exchanges GTP for GDP and dissociates (GTP and GDP separate)
3. GPCR-G protein complex disassembles, releasing the G protein a-subunit from BY complex
4. Ga-subunit binds to target enzyme (adenylyl cyclase), enhancing its activity
5. GTP hydrolyzed to GDP causing dissociation of a-subunit from target enzyme
6. a reassoociates with B Y - subunits and GPCR
-the blueprint of life
-contains instructions for making proteins within the cell
DNA
DNA is a polymer which is made up of
nucleotides
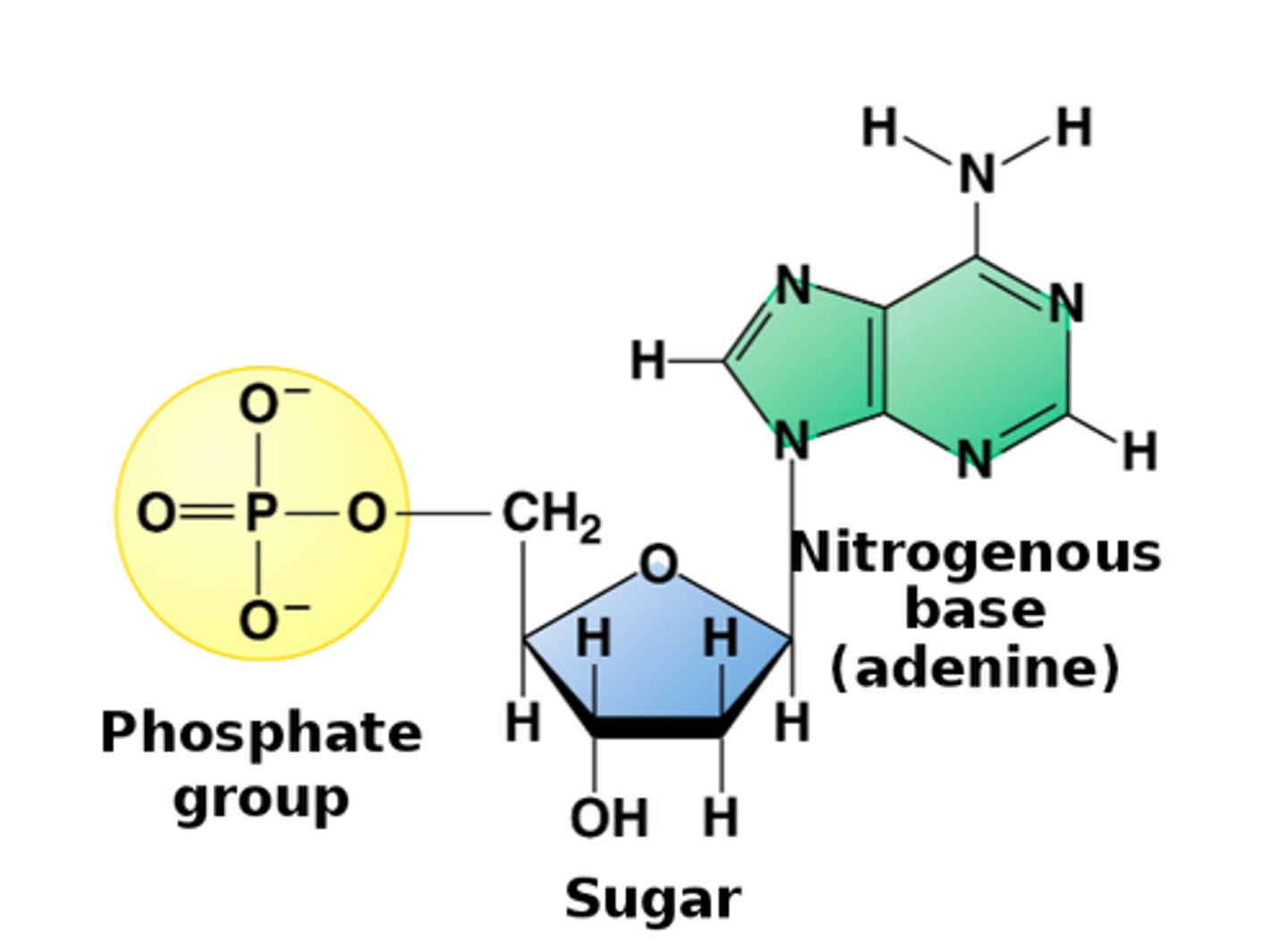
3 parts of nucleotide
nitrogen base
-purines, pyrimidines
pentose sugar
-ribose in RNA
-deoxyribose in DNA
phosphate (PO4) group
nitrogen base with double ring
purines
-adenine (A)
-guanine (G)
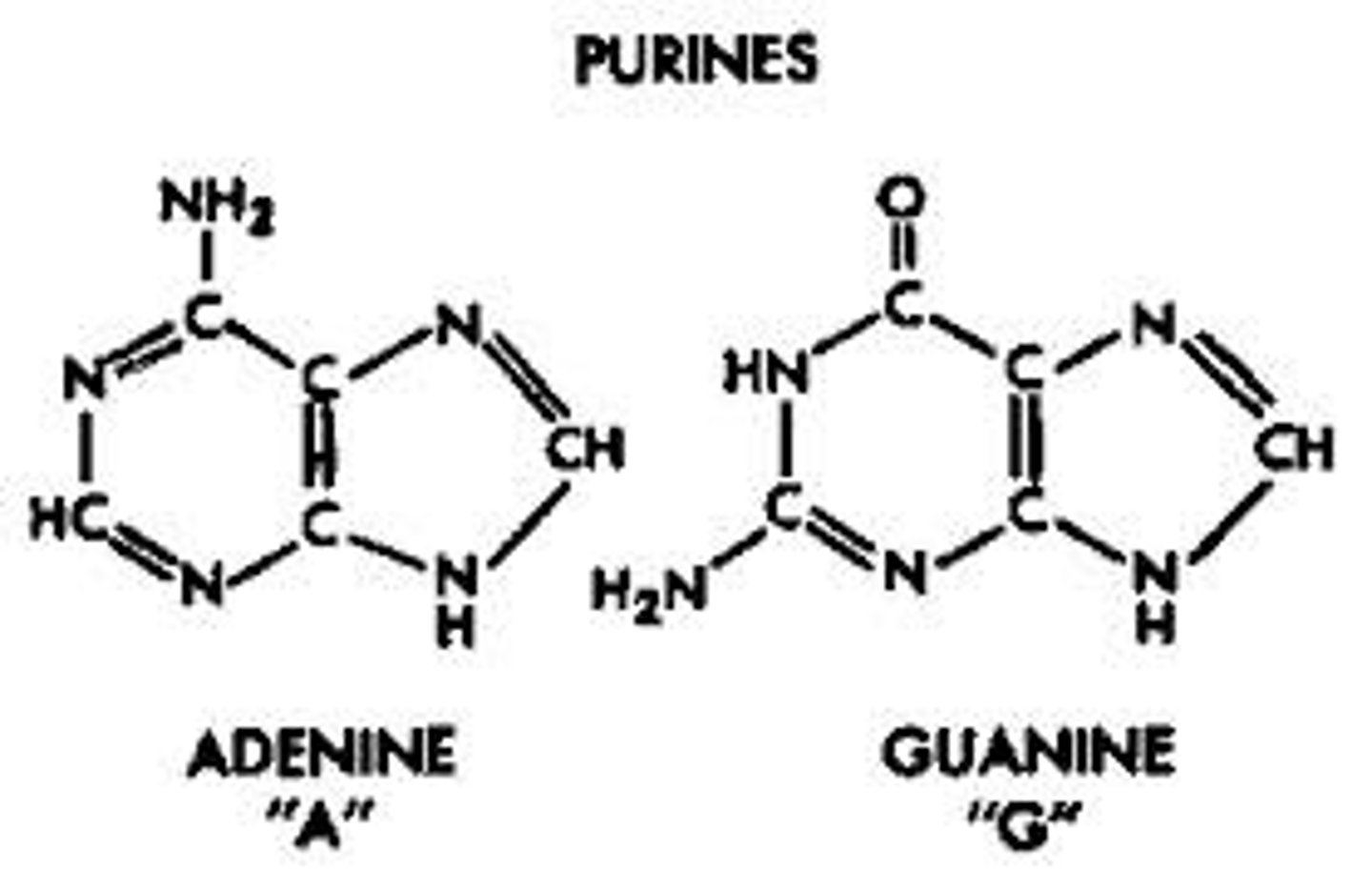
nitrogen base with single ring base
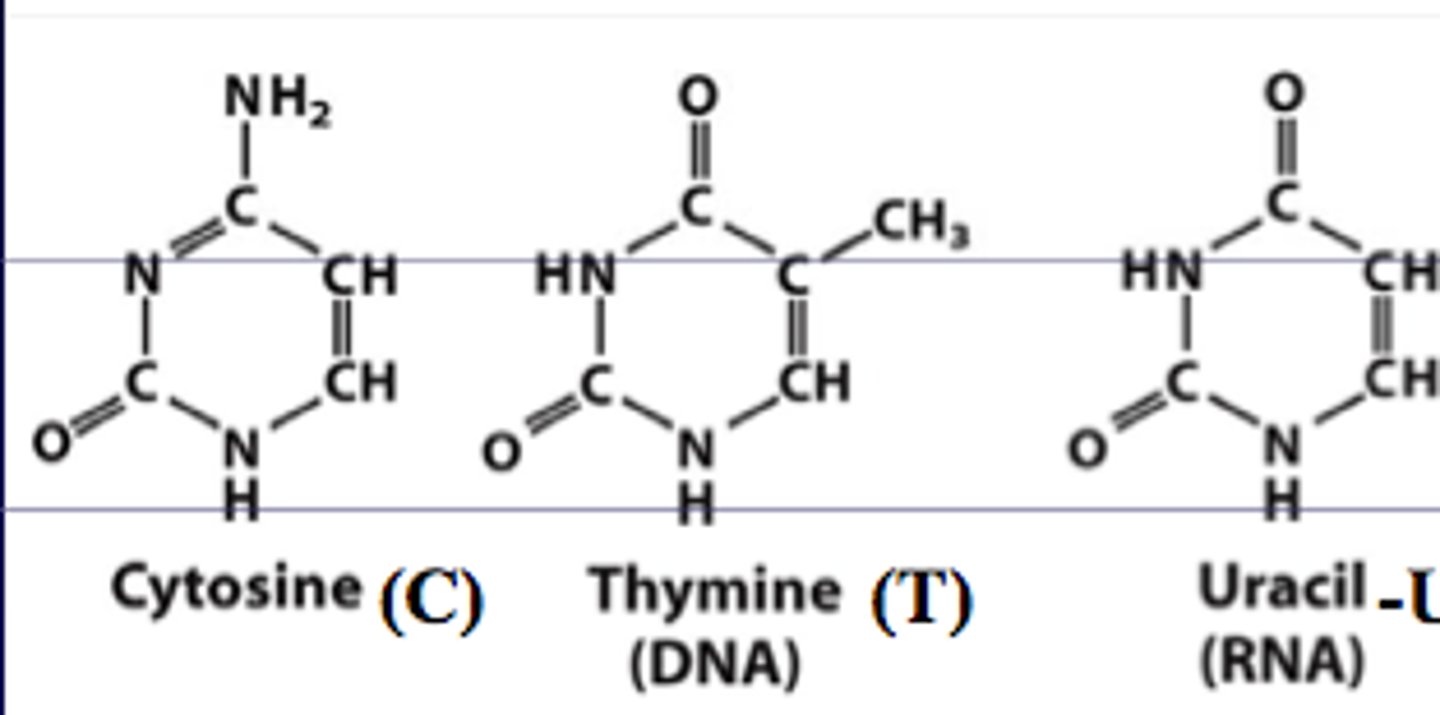
pyrimidines
-cytosine (C)
-thymine (T)
-uracil (U)
bond between nucleotides
hydrogen bond
purine::pyrimidine
Chargraff's Rule: Adenine always joins with
Thymine
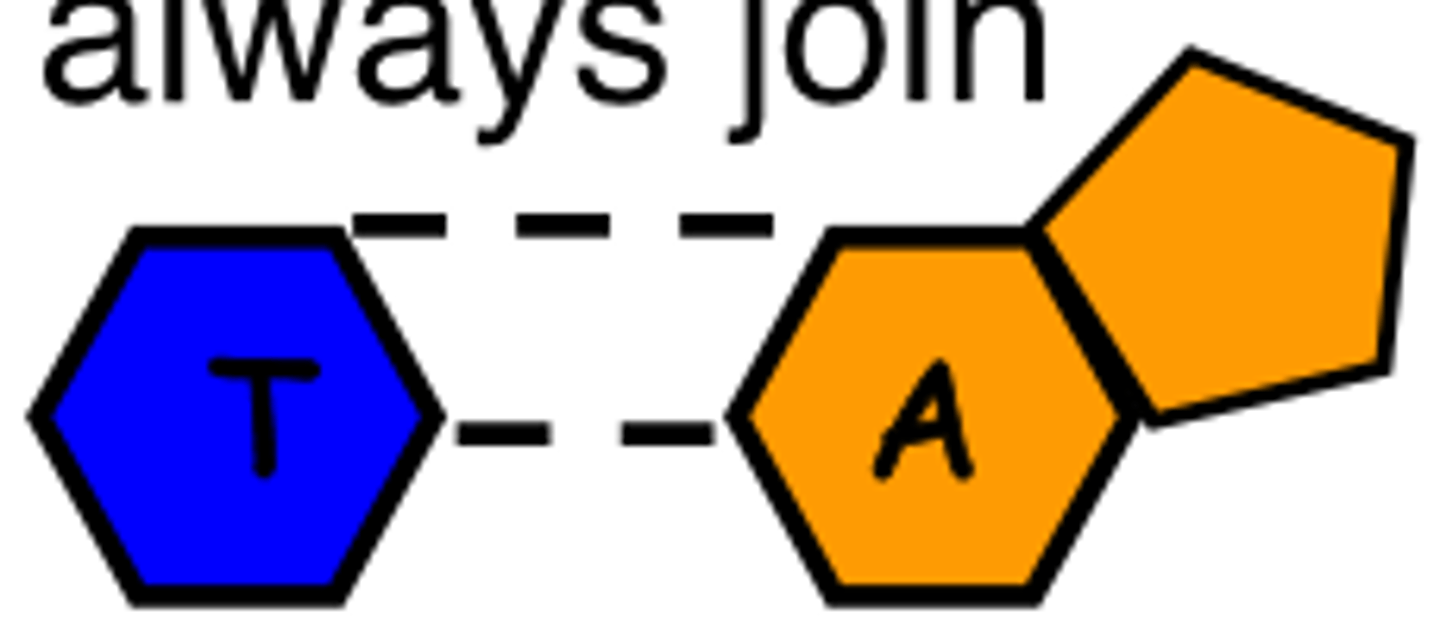
amount of H bonds between adenine and thymine
2
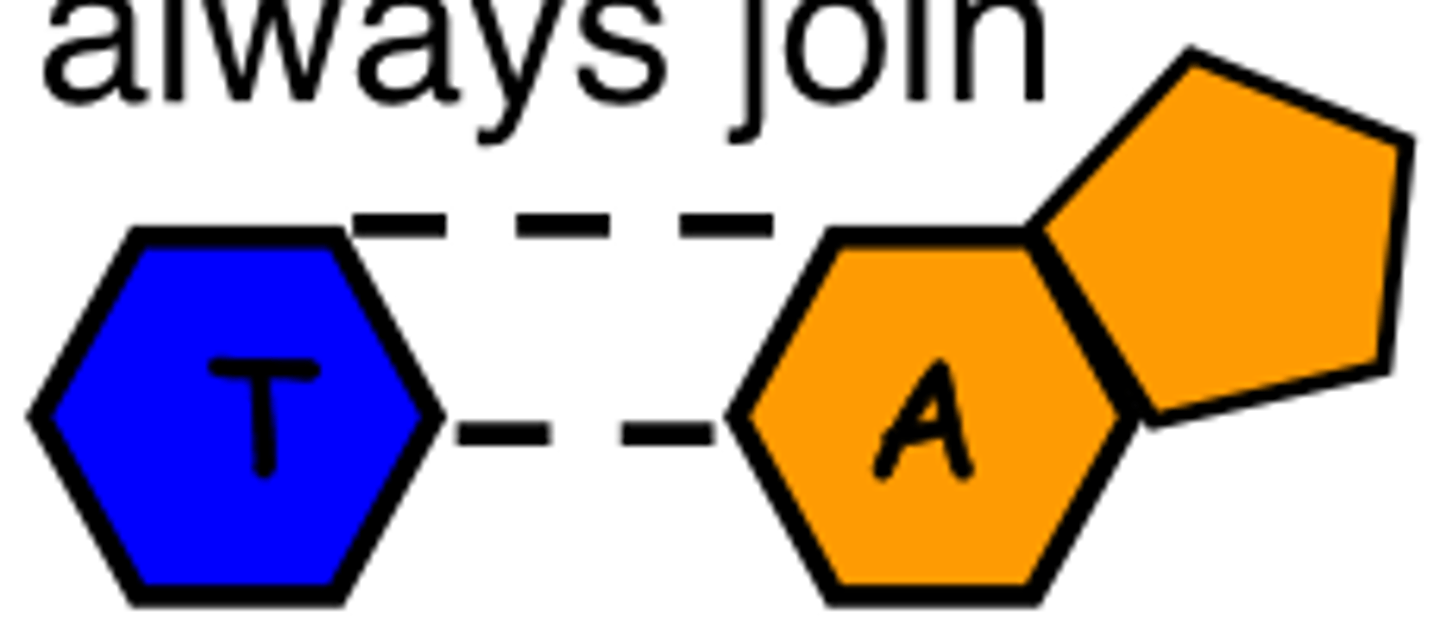
Chargraff's Rule: cytosine always bonds with
guanine
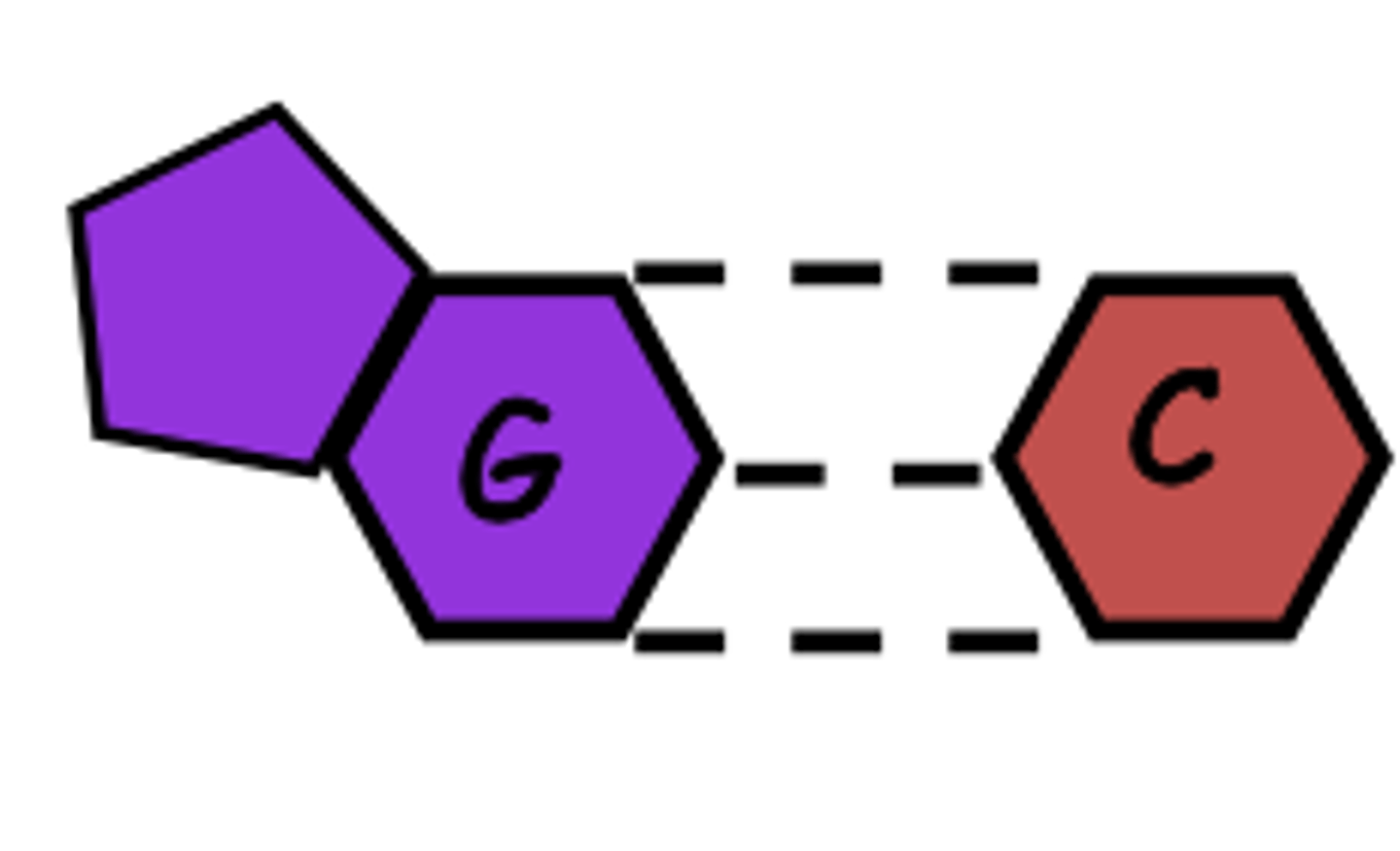
number of H bonds between cytosine and guanine
3
-stronger than adenine and thymine
one strand of DNA goes from 5' to 3' (sugars), the other strand is in what direction?
opposite
-antiparallel
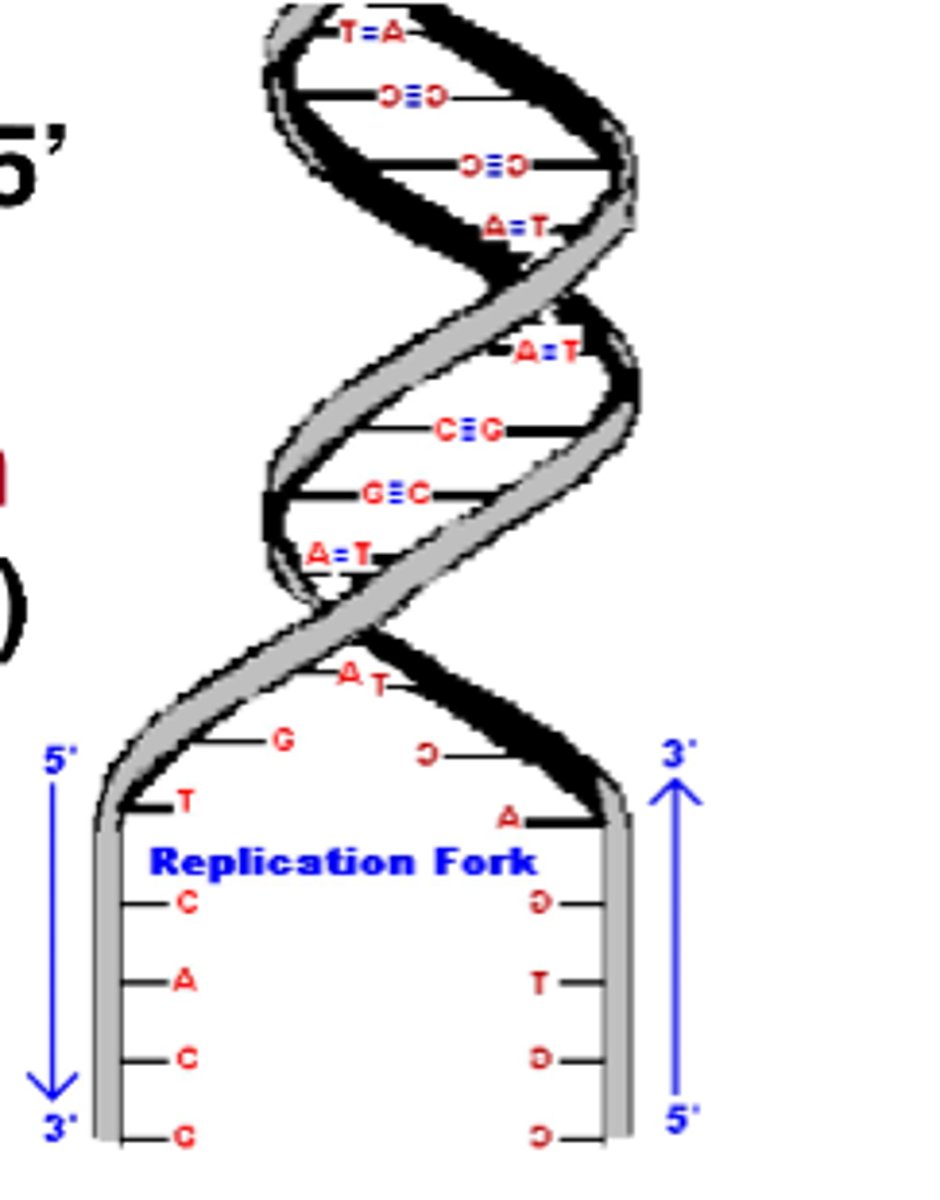
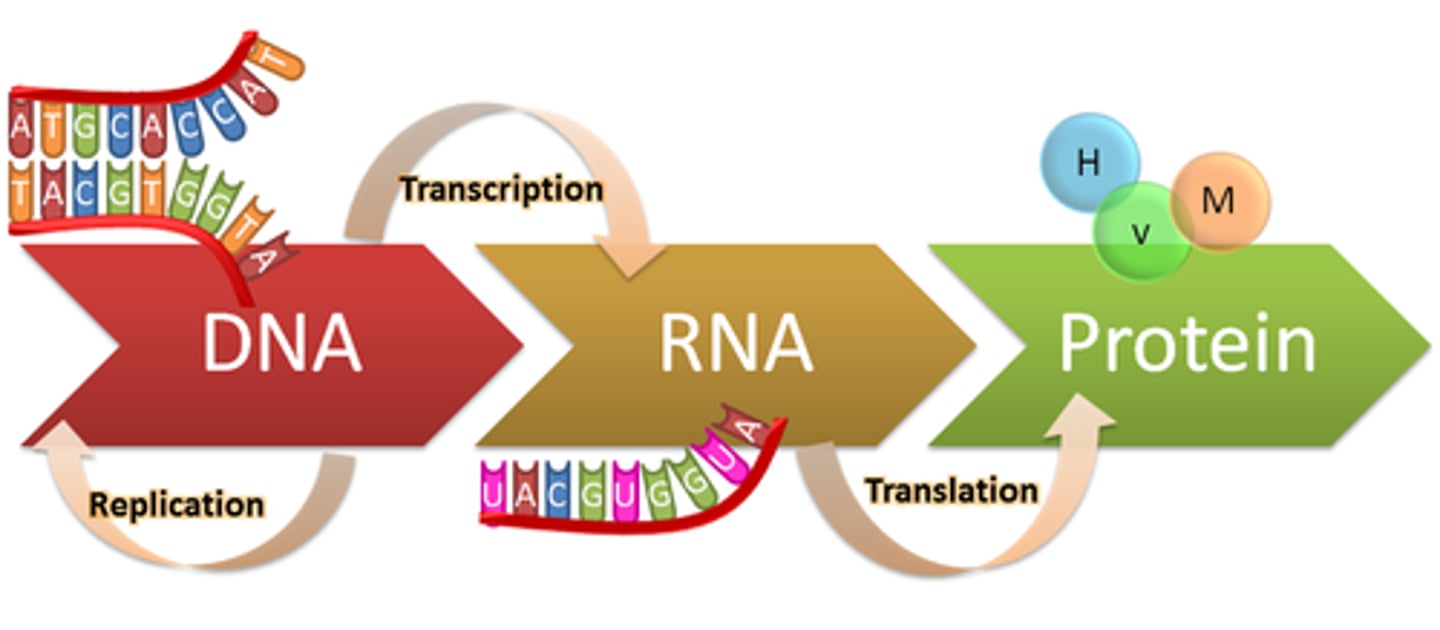
central dogma of molecular biology
DNA -> RNA -> Protein
why do we need DNA replication?
cell reproduction
-mitosis
gamete production
-meiosis
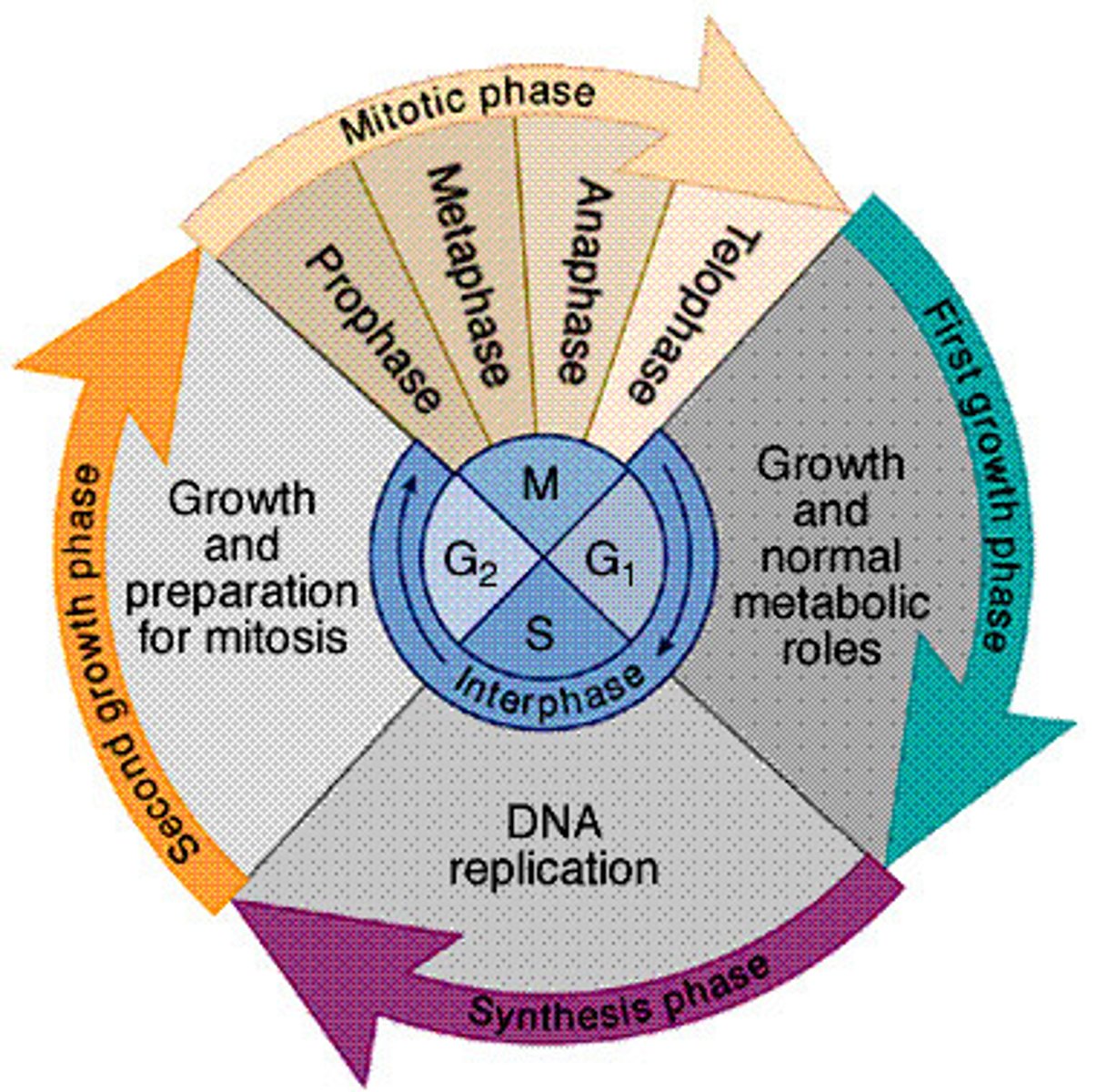
DNA replication takes place in the _______ phase
S
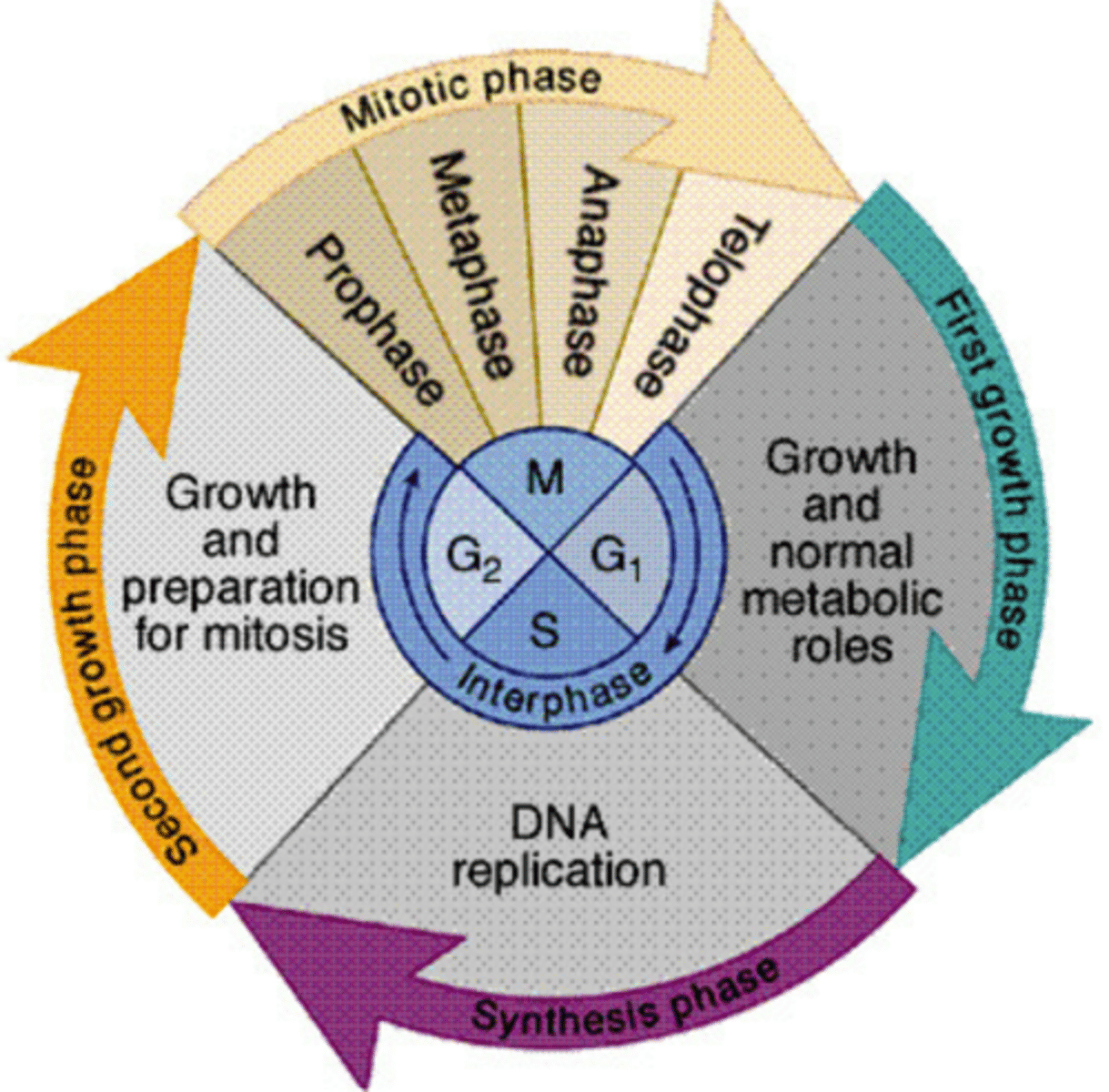
S phase during _________ of the cell cycle
interphase
DNA has to be _________ before a cell divides
copied
new cells will need __________ DNA strands
identical
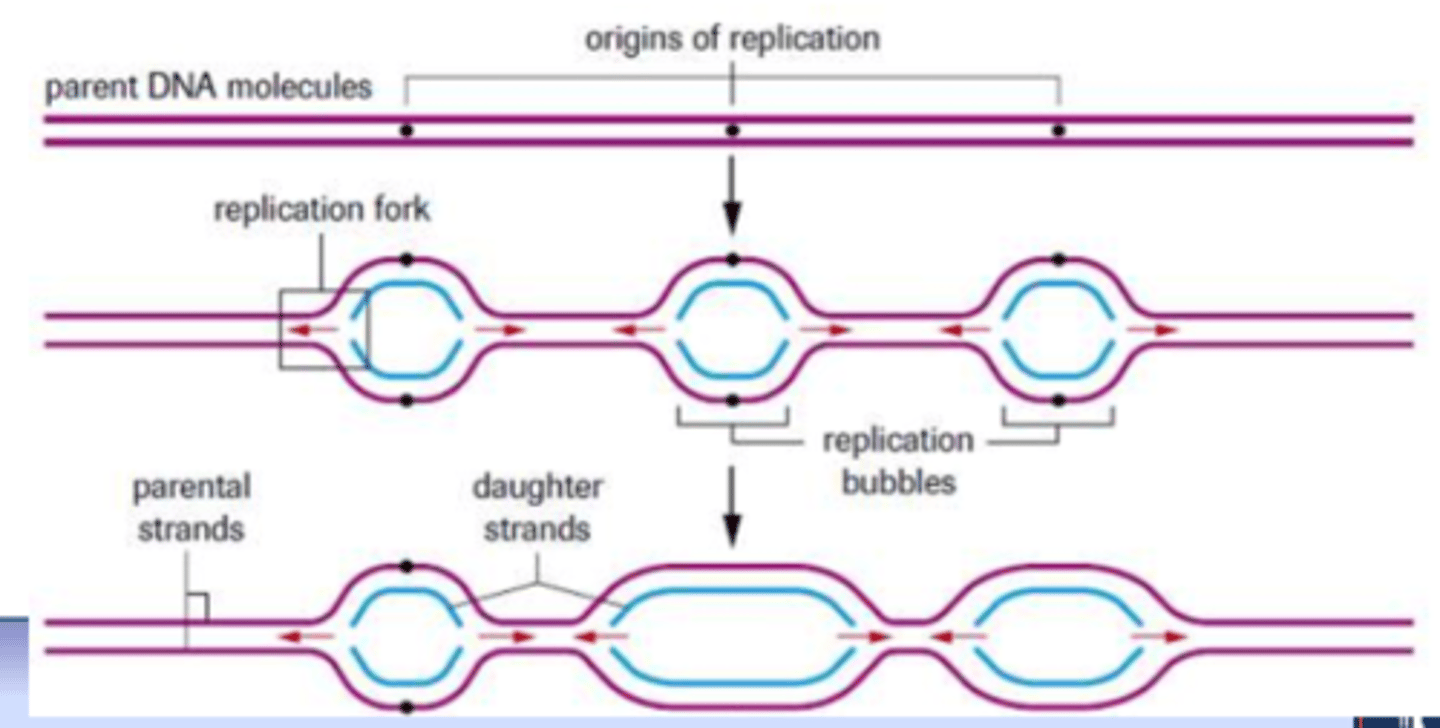
DNA replication begins at
origin(s) of replication
-in eukaryotes, hundreds/thousands of origin sites
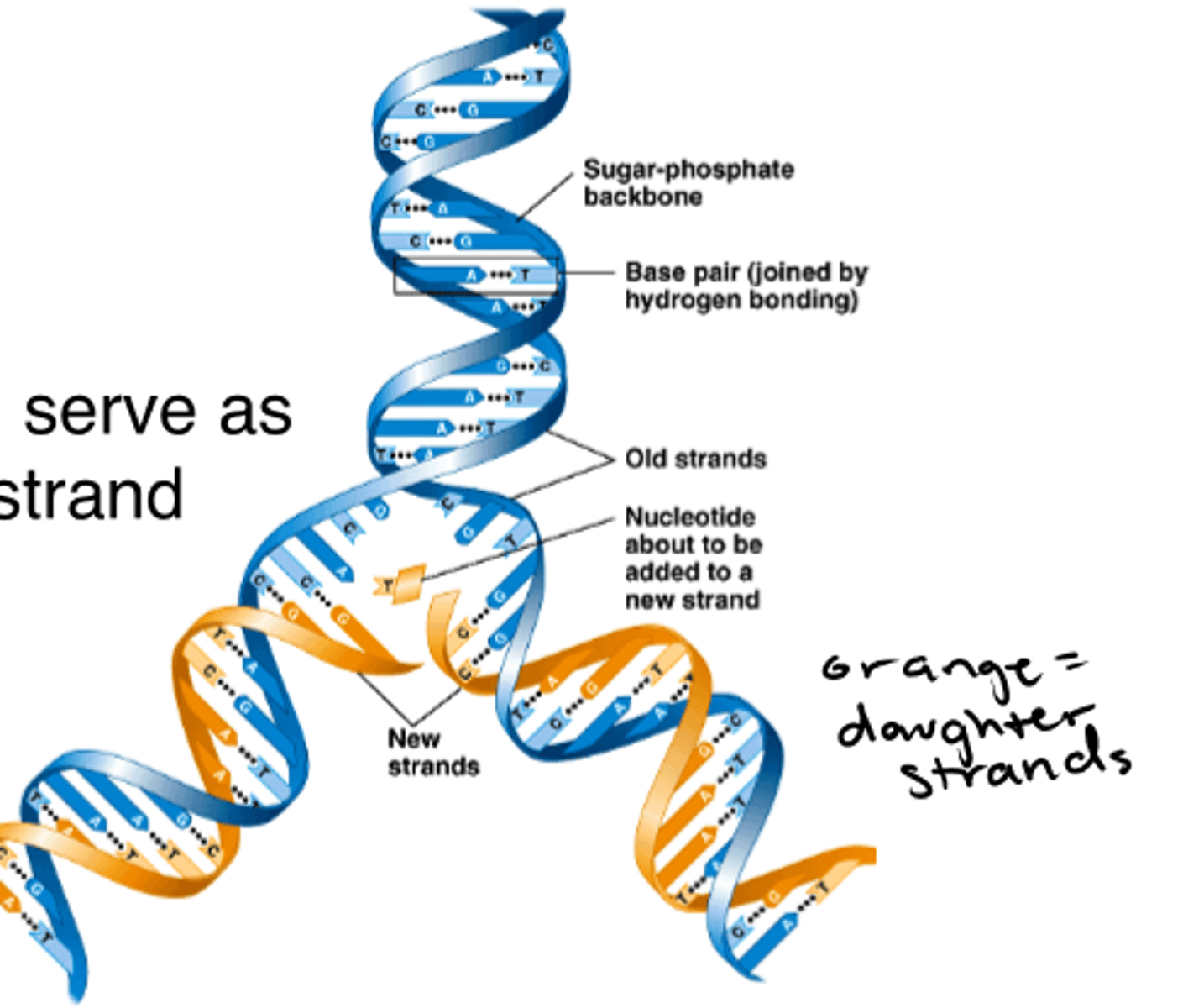
each DNA strand can serve as a _____________ for a new strand
template
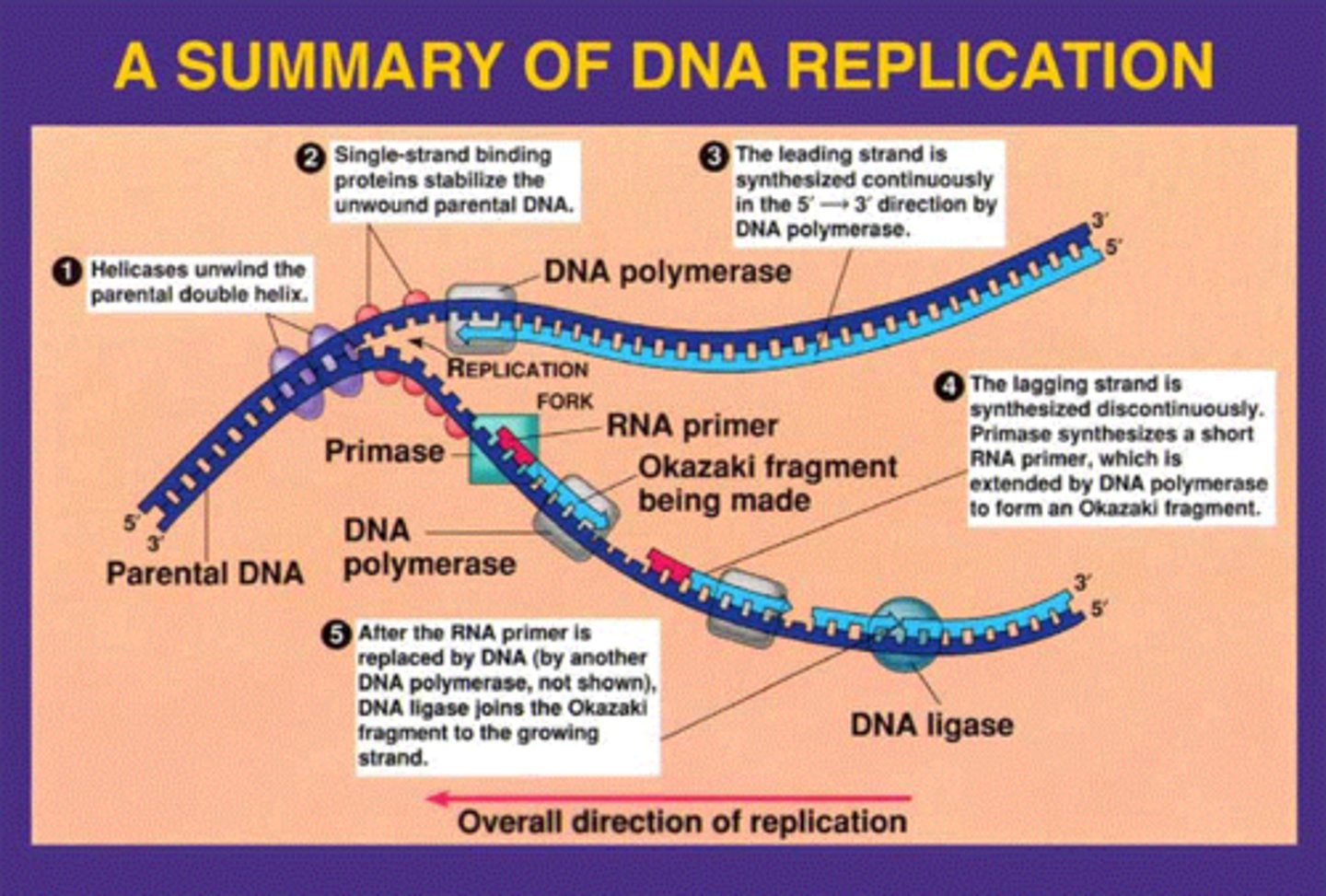
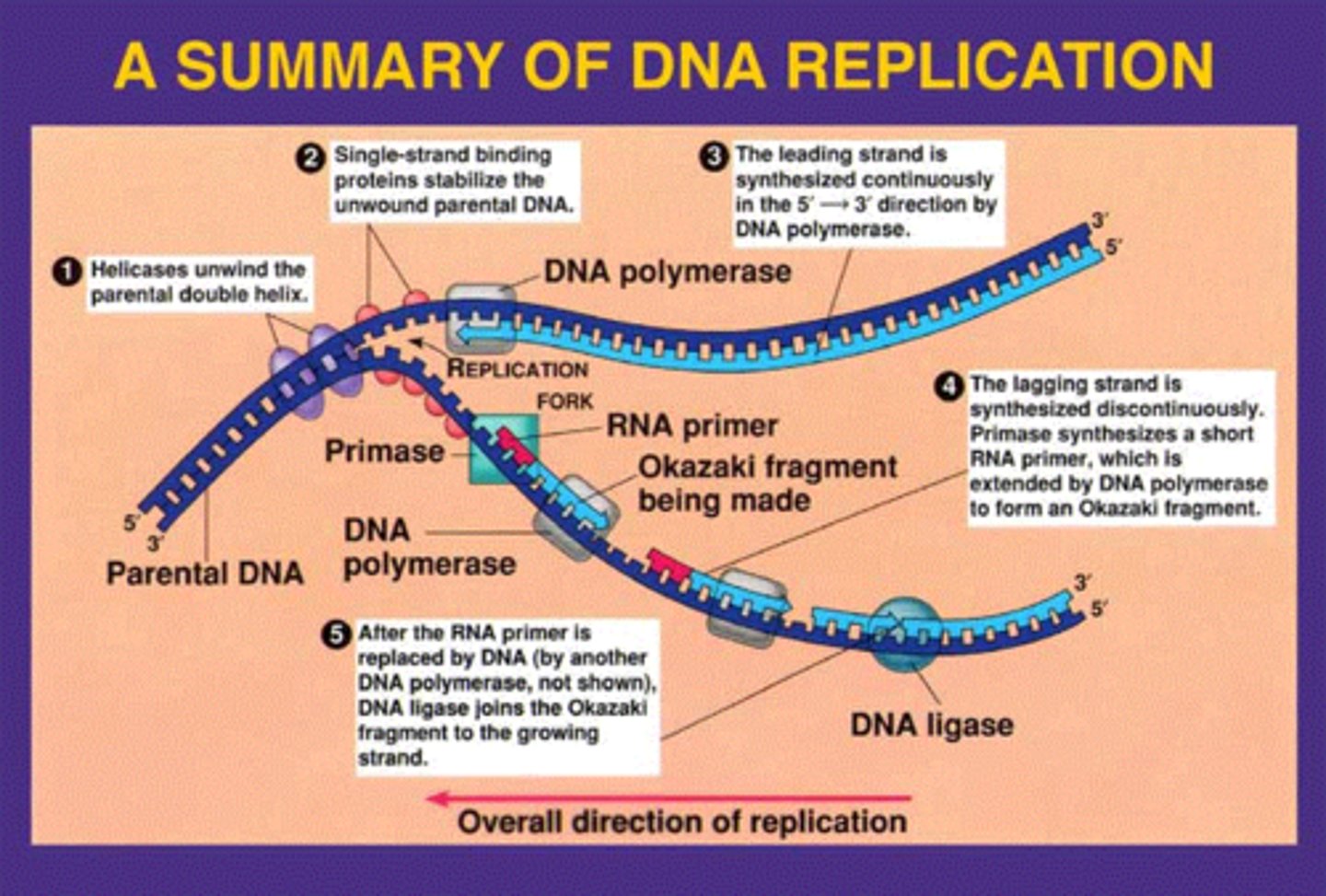
splitting of DNA during copying
replication forks
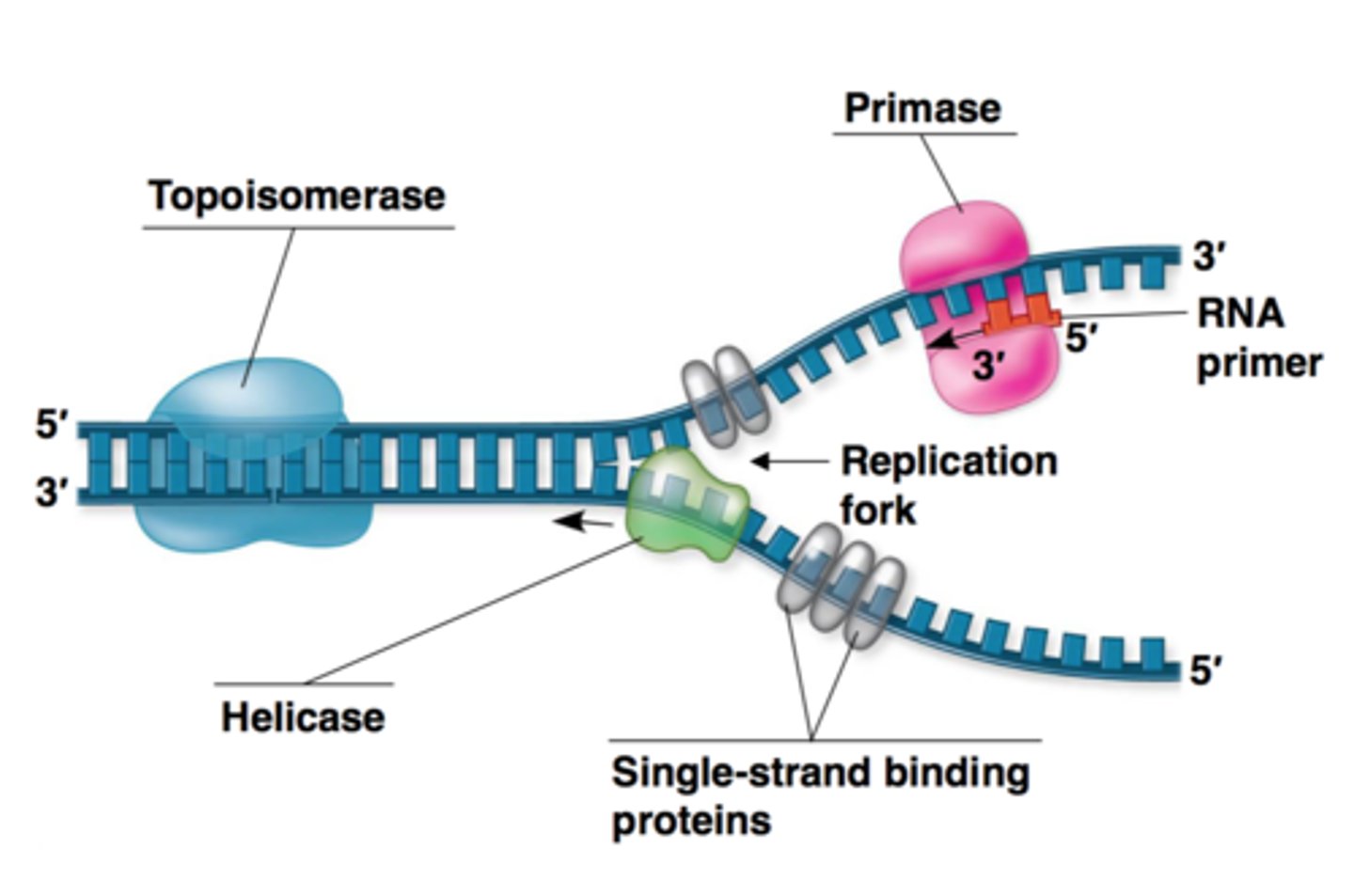
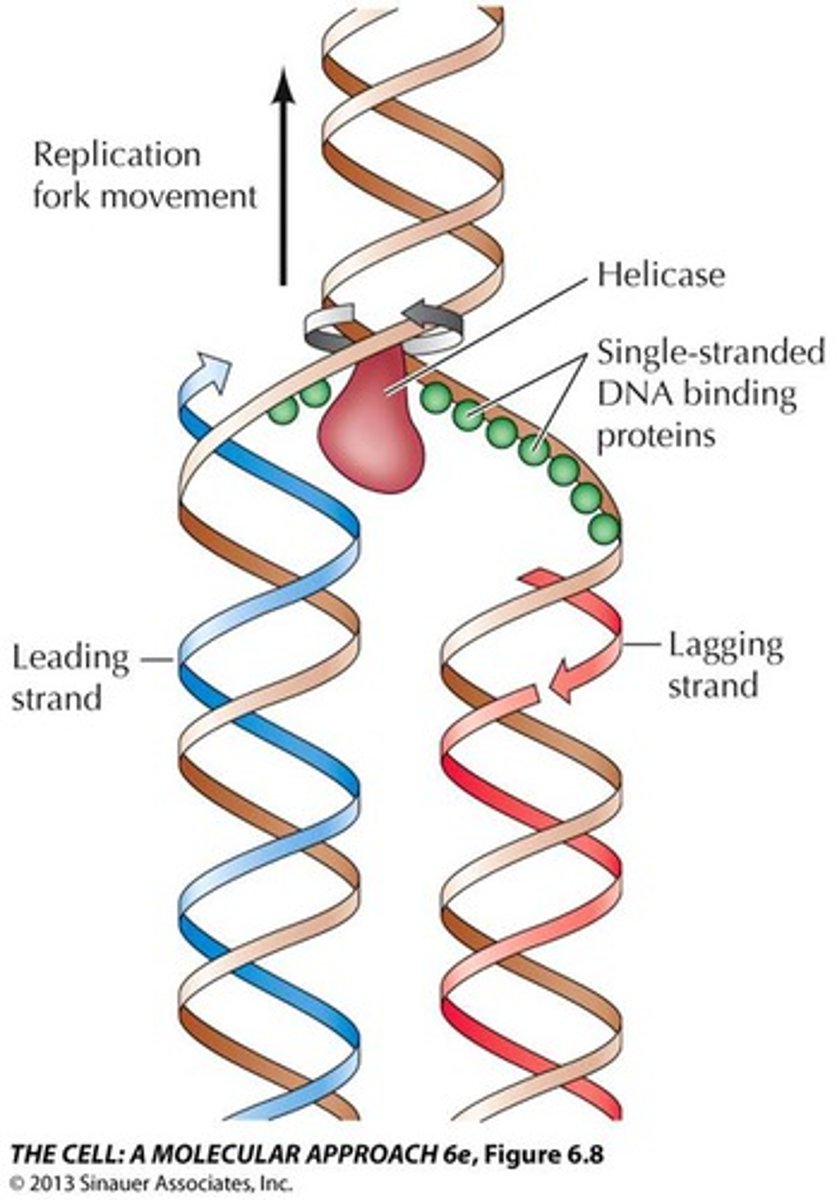
DNA strands are unwound by
DNA helicase
what prevent immediate reformation of the double helix?
single stranded binding proteins (SSB)
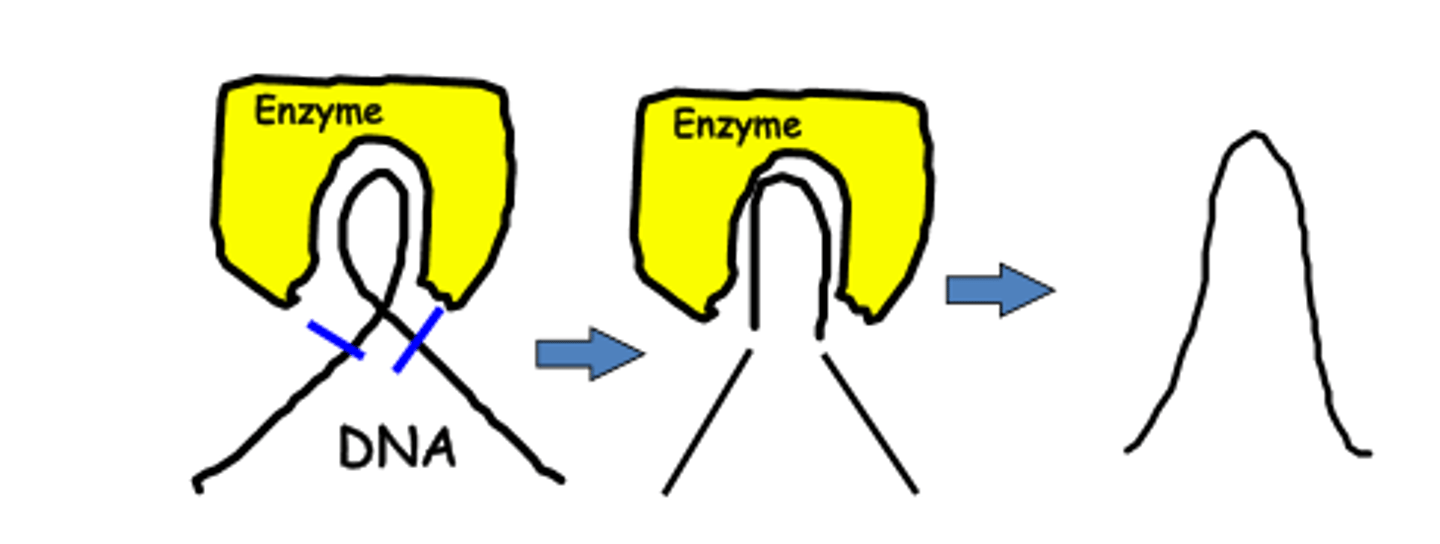
what "unties" the knots that form?
-attaches to the 2 forks of the bubble to relieve stress on DNA molecule as it separates
topoisomerase
synthesis RNA primer from a single template strand
-DNA polymerase can begin its chain after a few RNA nucleotides have been added
RNA Primase
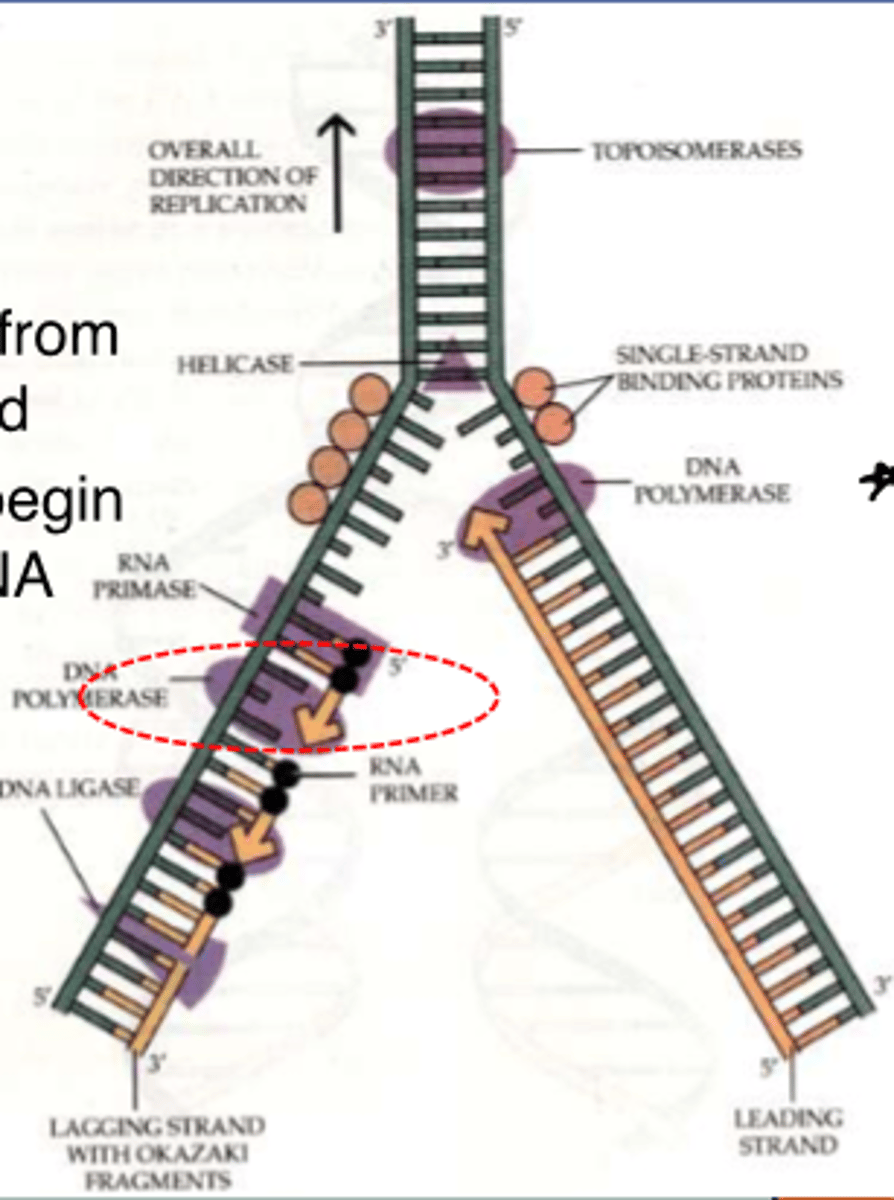
___________________ add nucleotides to the end of an existing chain
-it cannot initiate synthesis
-requires a "primer" synthesized by primase
-can add only to 3' end
DNA polymerase
new DNA strand can only elongate in what direction?
5' to 3'
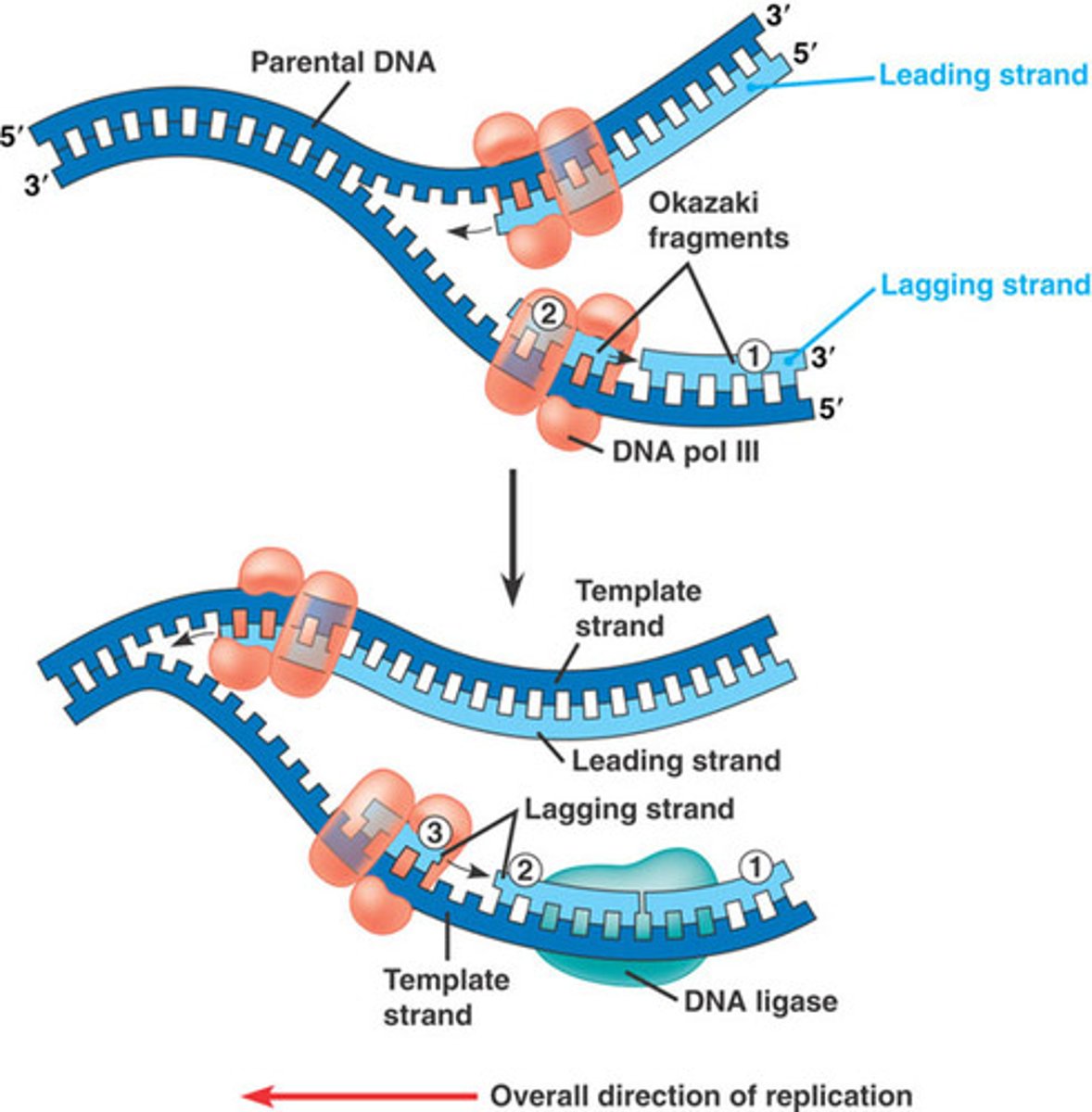
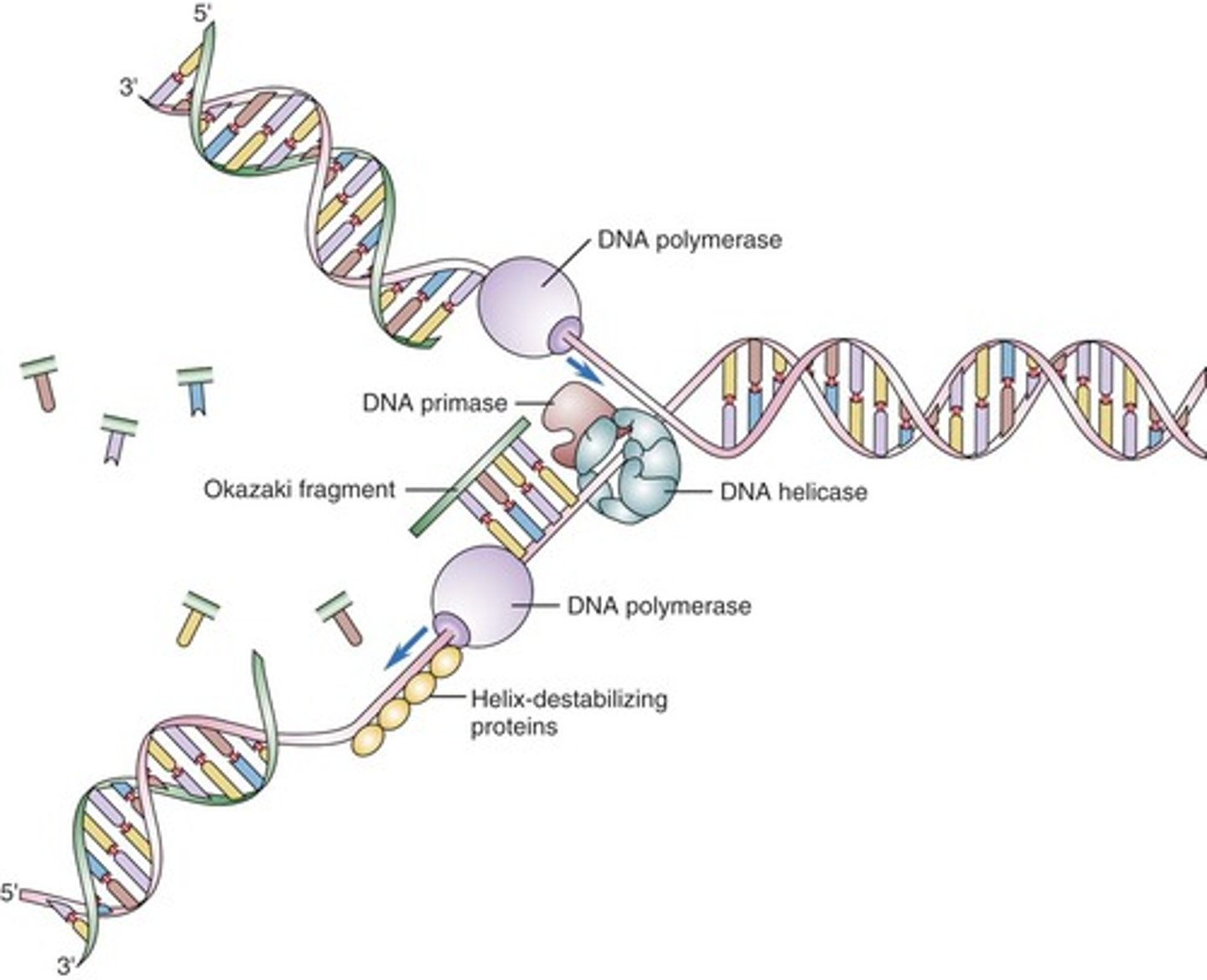
______________ is synthesized as a single strand from the point of origin toward the opening replication fork
leading strand
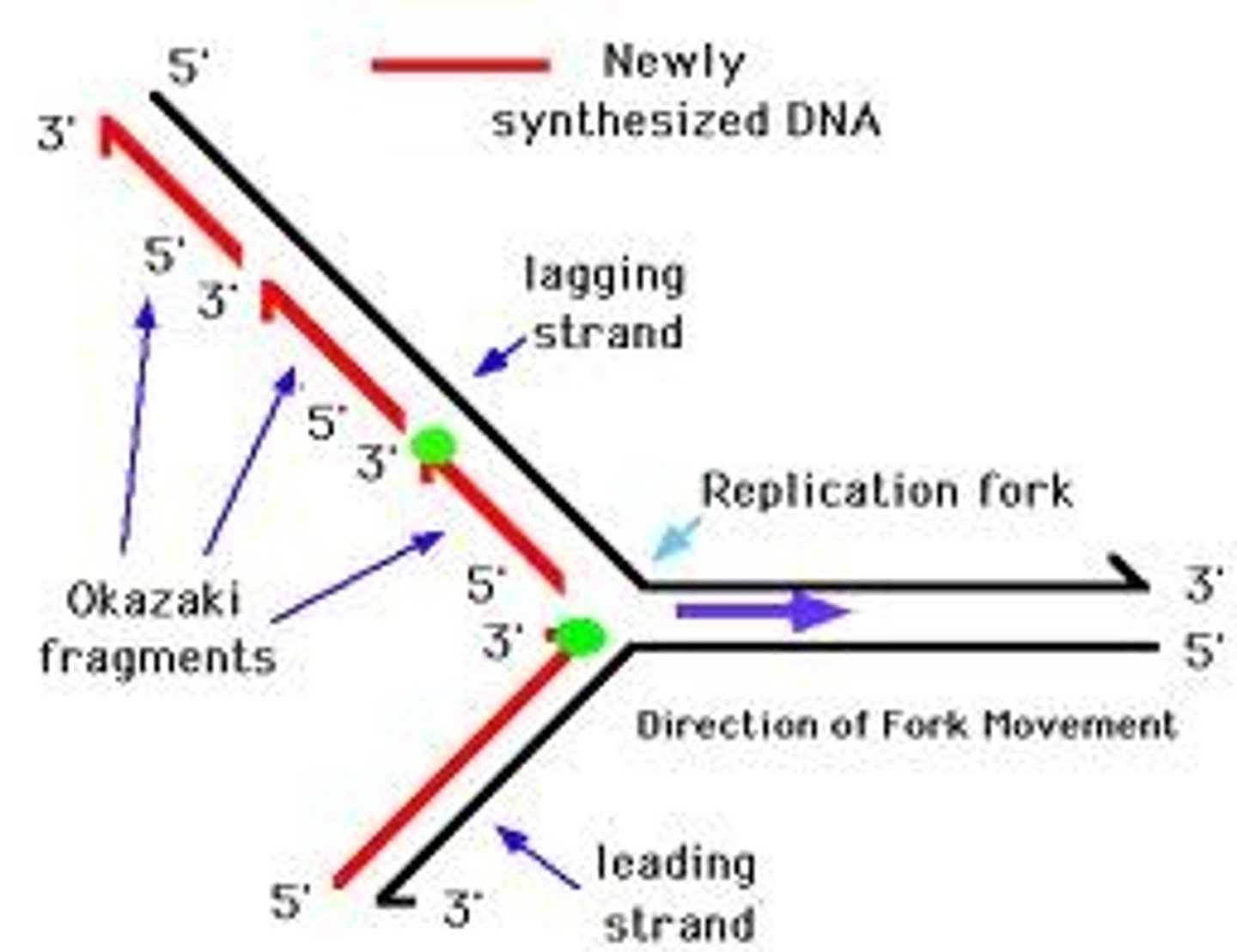
the _________________ is synthesized discontinuously against the overall direction of replication
-this strand is made of many short segments
-replicated from replication fork toward the origin
lagging strand
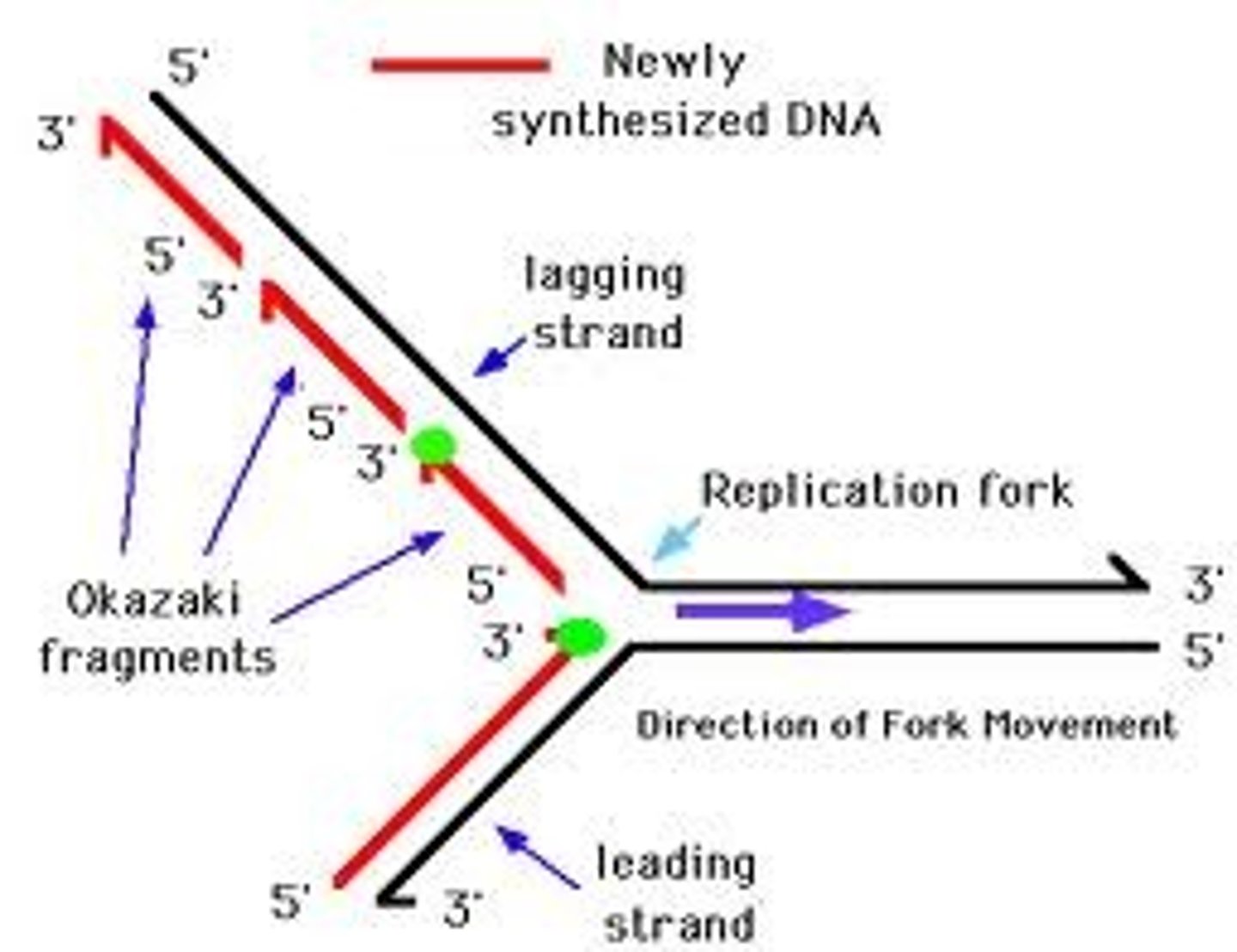
series of short segments on the lagging strand
-must be joined together by an enzyme
Okazaki Fragments
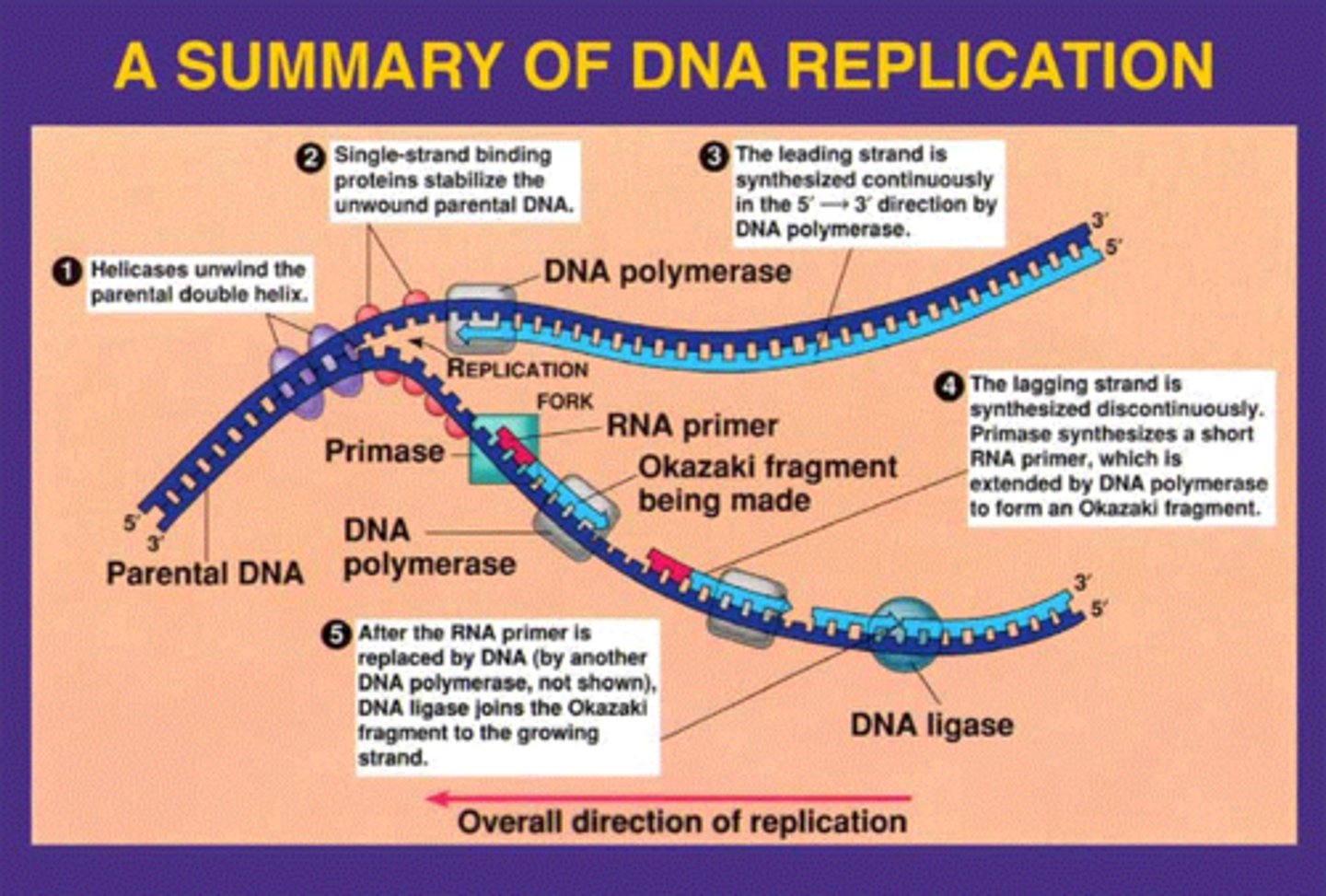
enzyme ___________ joins the Okazaki fragments together to make one strand
ligase