BIOL 350 - Principles of Genetics Exam 1
1/99
Earn XP
Description and Tags
KU Summer 2025
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
100 Terms
4 criteria for “genetic material”
Information
Transmission
Replication
Variation
Summary of Griffith’s Experiment on Genetic Transformation
Type S bacteria has a capsule that allows it to kill mice; type R bacteria does not have a capsule and cannot kill mice. Heat-killed type S bacteria and live type R bacteria together will kill a mouse because the type R uses the “transforming principle” to become type S (DNA)
Summary of Avery, MacLeod, & Mccarty’s experiment
Identified that DNA was the genetic material; cells with only DNA, not RNA or proteins, converted type R to type S. Additionally they added enzymes that destroy protein and RNA to ensure that DNA was the transforming principle
Summary of Hershey & Chase experiment
used microisotopes to distinguish DNA from proteins (phosphate for DNA and sulfur for proteins)
Which carbons of deoxyribose/ribose sugar does the dase, phosphate, and hydroxyl attach to?
base attaches to 1’
phosphate attaches to 5’
-OH hydroxyl attaches to 3’
What is a nucleotide made of? (3 parts)
Deoxyribose/ribose sugar
phosphate group
base
What is the structural difference between deoxyribose and ribose ?
ribose has a hydroxyl group on both the 2’ and 3’ carbon
deoxyribose has a hydroxyl group on only the 3’ carbon
Which bases are purines (how many rings) and which bases are pyrimidine (how many rings)
Purines (2 rings) are adenine and guanine and pyrimidines (1 ring) are thymine, uracil, and cytosine
Chagraff’s Rule
equal amounts thymine and adenine and equal amounts cytosine and guanine
3 stages of DNA replication
Initiation, Elongation, and Termination
Where is the prokaryotic origin of replication
oriC
3 types of DNA sequences at the oriC
AT-rich area
DnaA boxes
GATC methylation sites
What happens at the DnaA boxes?
DnaA proteins bind to DnaA boxes and bend DNA so it can be separated more easily
What is DnaB / helicase
unzips DNA for replication; travels 5’ to 3’
Function of GATC methylation sites
regulatie replication and ensure it only occurs once; daughter strand not immediately methylated, prevents second round of replication
Topoisomerase II / DNA Gyrase
travels ahead of replication fork and unwinds supercoiled DNA
Primase
synthesizes RNA primers and attaches them across the template strand
RNA Primer
allows DNA polymerase to attach nucleotides and create daughter strand of DNA
DNA polymerase III
adds nucleotides to make daughter strand
DNA polymerase I
removes RNA primers and puts DNA nucleotides in their place
DNA ligase
links okazaki fragments together
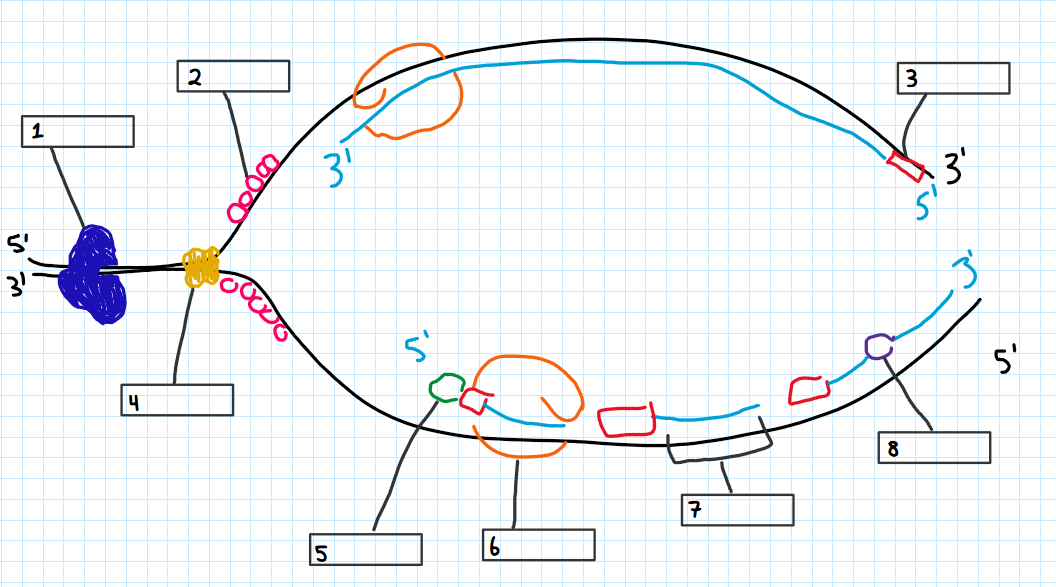
Name each labeled structure or protein of the replication bubble
Teloisomerase
single strand binding
RNA primer
DNA helicase
Primase
DNA polymerase III
okazaki fragment
DNA ligase
Leading strand
only one primer
Lagging Strand
multiple primers
What are okazaki fragments?
the fragments of replicated DNA that are attached to RNA primers in the lagging strand
What are the Ter sequences in prokaryotic DNA
termination sites opposite of oriC
What protein binds to the Ter sequences to stop the movement of the replication forks in prokaryotic DNA replication?
tus
What is termination utilization substance (tus)?
binds to Ter sequences to stop the replication fork in prokaryotic DNA replication
What are 3 fidelity mechanisms in DNA replication to minimize mistakes?
stability of correct base pairs
structure of the active site of DNA polymerase - distortion in shape due to mispairing prevents incorrect nucleotide fitting in active site
proofreading functions
What is the proofreading function of DNA polymerase?
3’ to 5’ exonuclease can digest the newly made strand and remove the mismatched nucleotide
Where is the origin of replication for eukaryotes?
ARS (autonomously replicating sequence)
How many origins of replication are present on a single chromosome of eukaryotic DNA? (1 or multilple?)
multiple
How does termination (DNA replication) work in eukaryotes?
The multiple replication bubbles meet each other and merge into completed chromosomes
Telomere
repetitive DNA sequences on 3’ overhang of linear chromosomes
Why can’t the 3’ ends of linear chromosomes be replicated?
DNA polymerase can only add DNA in 5’ to 3’ direction and cannot initiate synthesis. There is no room for an RNA primer at the 3’ end.
What does telomerase do?
adds RNA to the 3’ end of the template strand, allowing for an RNA primer to attach for the 3’ end DNA to be replicated as an okazaki fragment
What disease connects telomere length and aging?
Werner syndrome
Regulatory sequences (DNA transcription) binding site for ___ and function
binding site for regulatory proteins; influence the rate of transcription
Promoter (DNA transcription) binding site for ___ and function
binding site for RNA polymerase; signals start of transcription
Terminator (DNA transcription) function
signals the end of transcription
Start codon is usually ____ and/or ____
methionine or formylmethionine
Codon
3-nucleotide sequence of DNA that specifies a particular amino acid
Stop codon
specifes the end of polypeptide synthesis
Template strand
DNA strand that is “read” during transcriptionc
coding strand
RNA transcript is identical to it (T → U)
What do transcription factors do?
recognize promoter and regulator sequences to control transcription
3 stages of DNA transcription
initiation
elongation
termination
RNA polymerase
catalyzes the synthsis of RNA transcript
What groups compose the holoenzyme of RNA polymerase in prokaryotes like E.coli
core enzyme - 5 subunits
sigma factor - 1 subunit
Describe the initiation of prokaryotic DNA transcription, using the terms RNA polymerase, promoter, sigma factor, core enzyme, major groove, and open complex.
RNA polymerase moves along the DNA strand and scans until it encounters the promoter. Sigma factor recognizes this -35 to -10 reguin and binds to the major groove to create an open complex and then is released and the core enzyme.
2 mechanisms for termination of DNA transcription in E.coli
rho-dependent
rho-independent
steps of rho-dependent termination using terms rho recognition site (rut), rho protein, RNA polymerase, and stem loop
rho protein binds to the rho recognition site and moves in the 3’ direction. at the same time, RNA polymerase transcribes a region that forms a stem root, causing the polymerase to pause. during the pause, rho protein catches up to the open complex and separates the RNA-DNA hybrid.
two RNA sequences that facilitate rho independent termination
uracil rich sequence at 3’ end of the RNA
stem loop structure upstream of the uracil-rich region
Why is it that an enhancer can be thousands of nucleotides upstream of the promoter but is still able to facilitate RNA polymerase attachment?
enhancer can still be in close proximity to the promoter because of the folded and coiled nature of DNA
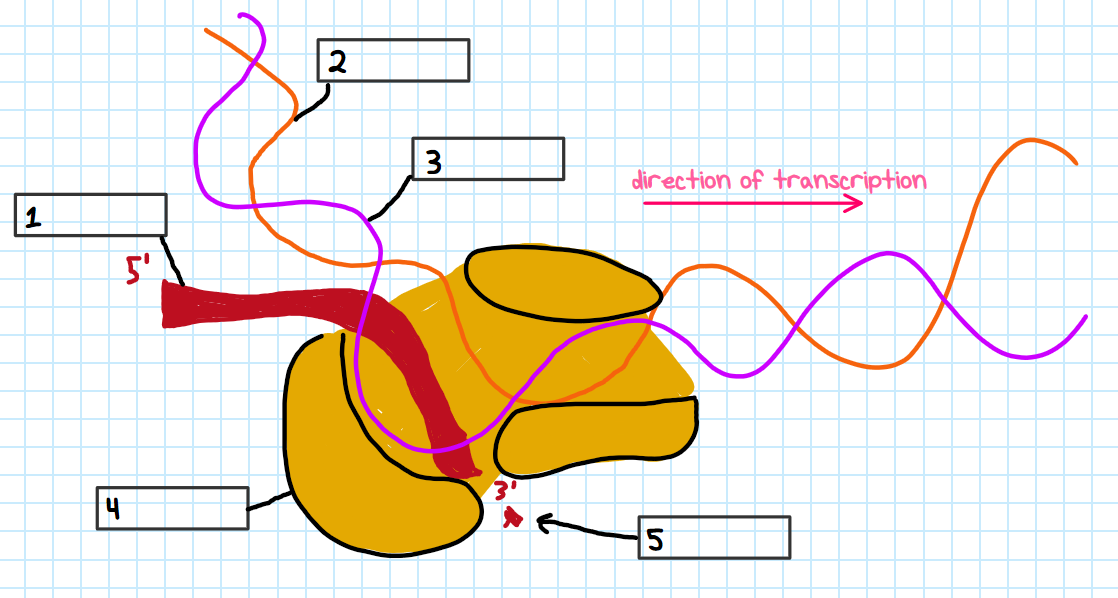
Label each number in the open complex figure
RNA transcript
coding strand
template strand
RNA polymerase
new nucleotide
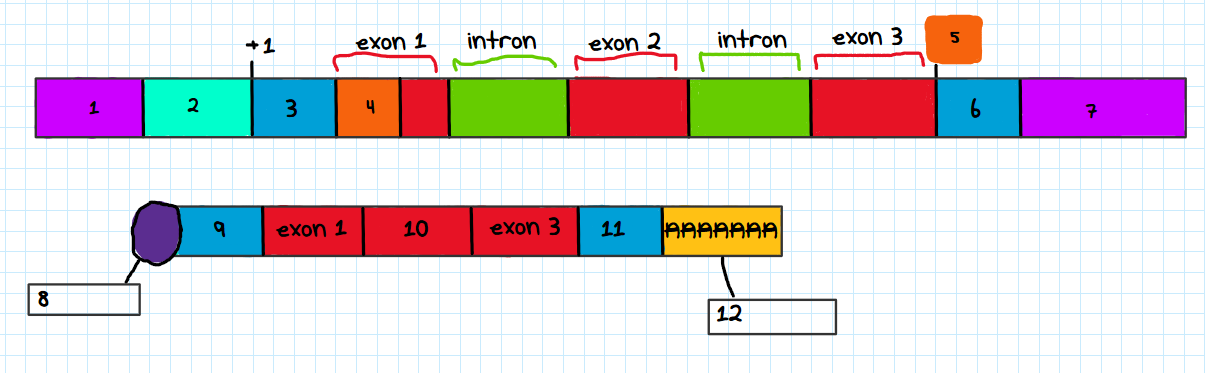
Label each number in the eukaryotic gene and processed mRNA transcript figure
enhancer
promoter
5’ UTR
start codon
stop codon
3’ UTR
termination site
5’ cap
5’ UTR
exon 2
3’ UTR
poly A tail
2 Functions of the 5’ cap on mRNA transcript
prevents degradation and aids in transportation of the transcript
Function of poly A tail in mRNA transcript
protects transcript at the 3’ end from being degraded
what binds to enhancers
transcription factors
function of enhancers
increase rate of transcription by helping recruit RNA polymerase
TFIID general transcription factor function
recognizes the TATA box
how does TFIID recognize TATA box?
TATA binding protein
what type of protein is GAL4
regulatory transcription factor
GAL4 aids transcription for what genes?
GAL enzymes
GAL 80 function
binds to GAL4 and inhibits transcrtiption
GAL3 function
bind to GAL80 in presence of galactose
the UAS (upstream activator sequence) is an ______?
enhancer
how does GAL4 initiate transcription?
recruits transcription complex
Why do you not want to transcribe genes when their encoded protein is not needed at the moment?
To save energy and efficiency
How does chromatin conformation affect transcription?
Closed conformation prevents transcription while open conformation DNA is accessible to transcription factors
Levels of Gene Regulation:
high level
binding to DNA
influence RNA pol ability to initiate transcrtiption
up- and down-regulation
chromatin conformation
DNA methylation
regulatory transcription factors
enhancers and silencers via activators and repressors
Acetylation of histone makes DNA ____
less tightly bound to the histone
Four levels of combinatorial control of gene transcrtiption
two or more regulatory transcription factors acting on same gene at once
modulation of activators and repressors
a. binding of small effector molecules
b. protein-protein interactions
c. covalent modifications
DNA methylation
What does DNA methylation do?
block transcription by preventing activators from binding
housekeeping genes
genes that are expressed consistently and constantly in most cell types
methylation ______ ______
inhibits transcription
gene regulation often occurs over ____ distances
long
acetylated DNA (is/is not) being actively transcribed
is
cis-acting elements
DNA sequences; exert effect over single gene
trans-acting elements
protein
What is promoter region on DNA called in 1. eukaryotes and 2. prokaryotes?
TATA box
-35 and -10 region
what recognizes promoter in prok. transcription?
sigma factor on RNA polymerase
what recognizes promoter in euk. transcription?
TFIID general transcription factor
What is the benefit of alternative splicing?
differential splicing allows different proteins created from same gene
What splices RNA transcript?
spliceosome
The two terminus names in a polypeptide chain
n terminus / amino
c terminus / carboxyl
The first amino acid in a chain is attached to what terminus?
n terminus
what is a charged tRNA
tRNA with an animo acid attached
what enzyme charges tRNA?
aminoacyl-tRNA synthetase
2 ribosome subunits:
large subunit
small subunit
translation initiation in 1. prokaryotes and 2. eukaryotes
16s rRNA recognizes shine delgarno sequence in 5’ UTR
small subunit attaches to 5’ cap and scans for start codon
Why do stop codons terminate translation?
No tRNA can recognize the codon. No new animo acids can be added to the chain
What recognizes the stop codon(s) in translation?
release factors that mimic tRNA shape
How can antibiotics target bacterial RNA translation but not harm human RNA translation?
bacterial ribosome is smaller than eukaryote
What does a repressor protein do?
blocks transcription when bound to DNA
What does an activator protein do?
starts transcription when bound to DNA
What does an inducer molecule do in gene regulation?
binds to a protein to induce a conformational change in shape
2 functions of inducer molecules in gene regulation
facilitate or inhibit the ability of a protein to bind to DNA
What is an operon?
regulatory unit of DNA encoding multiple genes under one promoter